Figures & data
Table 1. Primers used in this study.
Table 2. siRnas used in this study.
Figure 1. Screening the effects of hnRnps on SHVV replication. (A) CCO cells were transfected with siRnas targeting each of the twelve hnRnps, and the total RNAs were extracted at 24 h post transfection. The mRNA level of the hnRnps was measured using qRT-PCR, β-actin was used as the internal control. (B) CCO cells were transfected with siRnas targeting each of the twelve hnRnps, followed by SHVV infection. The total RNAs were extracted at 24 h post of SHVV infection, and the viral G mRNA levels in cells were measured using qRT-PCR, β-actin was used as the internal control. (C-D) CCO cells were transfected with siNC or sihnRNPA1, followed by SHVV infection. The G protein levels in cells were measured using Western blotting with β-actin as the internal control, while the viral titre in supernatants was measured using TCID50. All the data are performed in triplicate (mean ± sd). The * and ** indicate statistically significant differences (*p < 0.05; **p < 0.01).
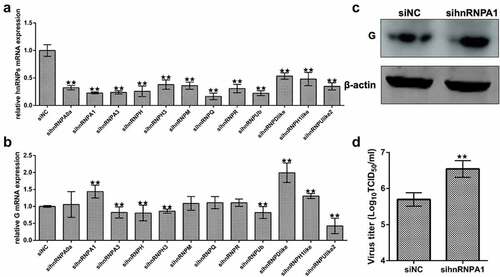
Figure 2. Effect of overexpression of hnRNPA1 on SHVV replication. (A and B) CCO cells were transfected with pFlag-hnRNPA1 or p3×flag-CMV-14 (control). The mRNA and protein levels of hnRNPA1 were measured using qRT-PCR and Western blotting, and β-actin was used as the internal control. (C-E) CCO cells were transfected with pFlag-hnRNPA1 or p3×flag-CMV-14, followed by SHVV infection. The viral G mRNA and protein levels in cells were measured using qRT-PCR and Western blotting, and β-actin was used as the internal control, while the viral titre in supernatants was measured using TCID50. All the data are performed in triplicate (mean ± sd). The * and ** indicate statistically significant differences (*p < 0.05; **p < 0.01).
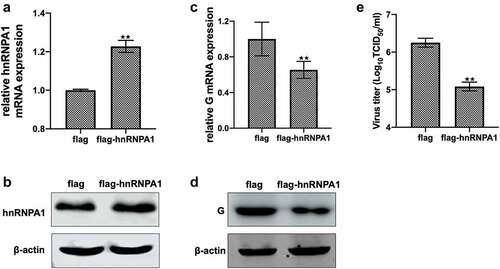
Figure 3. Expression of hnRNPA1 during SHVV infection. (A) CCO cells were infected with SHVV, and the cells were harvested at 0, 3, 6, 12, and 24 h. The mRNA levels of hnRNPA1 were measured using qRT-PCR, and β-actin was used as the internal control. (B) the protein levels of hnRNPA1 and SHVV G were measured using Western blotting, and β-actin was used as the internal control. (C-D) the plasmids expressing N, P, M, G, or L were transfected into CCO cells, the empty vector pCDNA3.1 was used as control. The mRNA and protein levels of hnRNPA1 were measured using qRT-PCR, and β-actin was used as the internal control. All the data are performed in triplicate (mean ± sd). The * and ** indicate statistically significant differences (*p < 0.05; **p < 0.01).
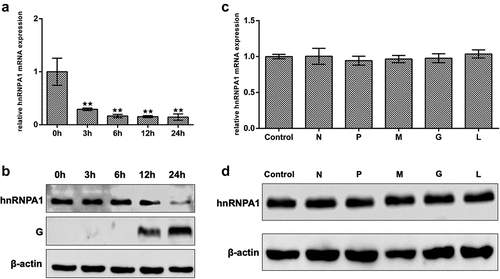
Figure 4. Subcellular localization of hnRNPA1 during SHVV infection. (A) CCO cells were infected with SHVV, and the cells were harvested at 0, 3, 6, 12, and 24 h post infection. The localization of hnRNPA1 was measured by immunofluorescence assay. (B) CCO cells were infected with SHVV, and the cytoplasmic and nuclear components were extracted at 0, 3, 6, 12, and 24 h post infection. Samples were detected by Western blotting with anti-hnRNPA1, anti-histone H3, and anti-β-actin antibodies. (C) the plasmids expressing N, P, M, G, or L were transfected into CCO cells, with the empty vector pCDNA3.1 used as control. The cytoplasmic and nuclear components were extracted, and the samples were detected by Western blotting with antibodies against hnRNPA1, histone H3, and β-actin.
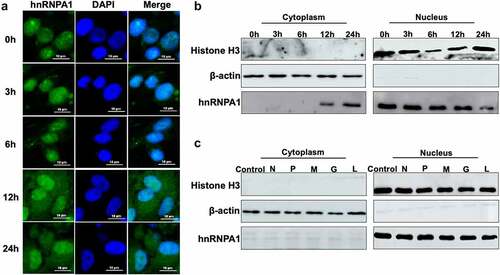
Figure 5. hnRNPA1 interacts with SHVV P protein. (A) CCO cells were transfected with pFlag-hnRNPA1 or p3×flag-CMV-14 (control), followed by SHVV infection. The whole-cell lysates were obtained at 24 h post infection and immunoprecipitated with anti-Flag antibody. The anti-N, anti-P, anti-L, and anti-Flag antibodies were used for Western blotting. (B-D) 293T cells were transfected with pFlag-hnRNPA1, together with the plasmids expressing His-N, His-P, or L. The whole-cell lysates were obtained at 24 h post transfection and immunoprecipitated with anti-Flag antibody. The anti-L, anti-His, and anti-Flag antibodies were used for Western blotting.
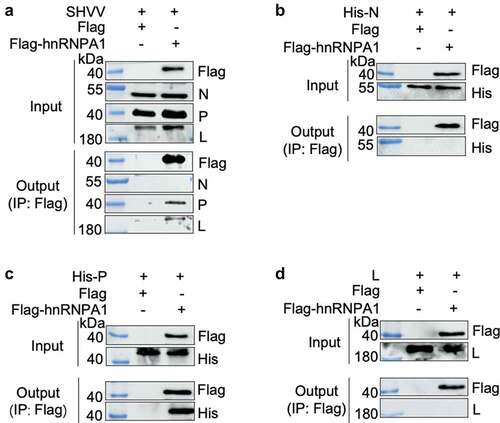
Figure 6. hnRNPA1 interacts with SHVV P through its Gly-rich region. (A) Sequence of hnRNPA1 from Ictalurus punctaus was compared with hnRNPA1 from Homo sapiens. Regions of hnRNPA1 was marked with different colours. (B) a series of hnRNPA1 segment plasmids expressing EGFP-fused RRM region, Gly-rich region, or M9 region were constructed. (C) 293T cells were transfected with plasmid pHis-P, together with plasmids expressing EGFP-fused RRM region, glycine-rich region, or M9 region of hnRNPA1. The whole-cell lysates were obtained at 24 h post transfection and immunoprecipitated with anti-EGFP antibody. The anti-P and anti-EGFP antibodies were used for Western blotting.
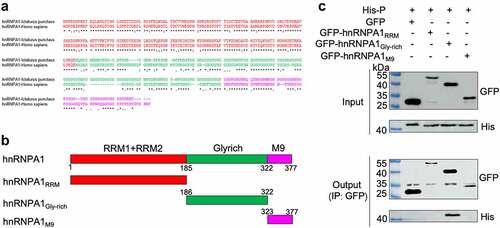
Figure 7. hnRNPA1 disrupts the viral P-N interaction. (A-C) 293T cells were transfected with pMyc-P and pFlag-hnRNPA1 or p3×flag-CMV-14 (control), together with plasmids expressing His-N, His-P, or L. The whole-cell lysates were obtained at 24 h post transfection and were immunoprecipitated with anti-Myc antibody. The anti-Myc, anti-His, anti-Flag and anti-L antibodies were used for Western blotting.
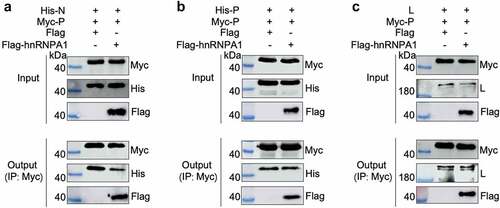
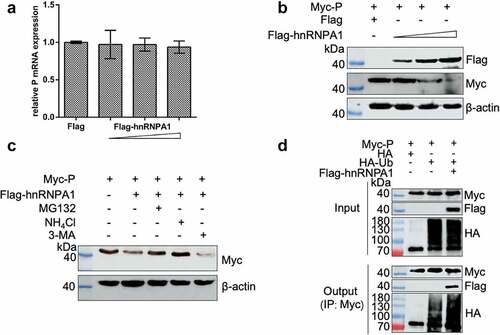
Figure 8. hnRNPA1 degrades SHVV P through proteasomal and lysosomal pathways. (A-B) 293T cells were transfected with pMyc-P, together with p3×flag-CMV-14 (control) or different doses of pFlag-hnRNPA1. The mRNA and protein levels of hnRNPA1 were measured using qRT-PCR and Western blotting with β-actin as the internal control. (C) 293T cells were transfected with pMyc-P together with pFlag-hnRNPA1, and the cells were treated with MG132 (5 µm), NH4Cl (15 µm), or 3-MA (60 µm). The cells were harvested at 24 h post transfection. The hnRNPA1 protein was determined by Western blotting, β-actin was used as the internal control. (D) 293T cells were cotransfected with pMyc-P and pHA-Ub, together with or without pFlag-hnRNPA1. The cells were collected at 24 h post transfection, and Co-IP assay was carried out with anti-Myc antibody. The anti-Myc, anti-Flag, and anti-HA antibodies were used for Western blotting.