Figures & data
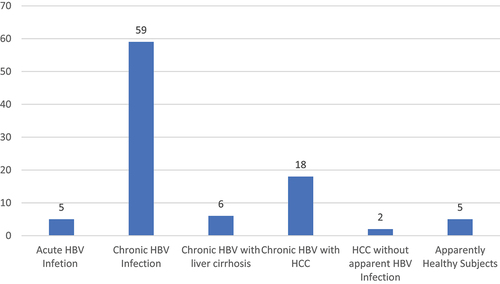
Table 1. Socio-demographic characteristics and HBV serological profiles of study participants.
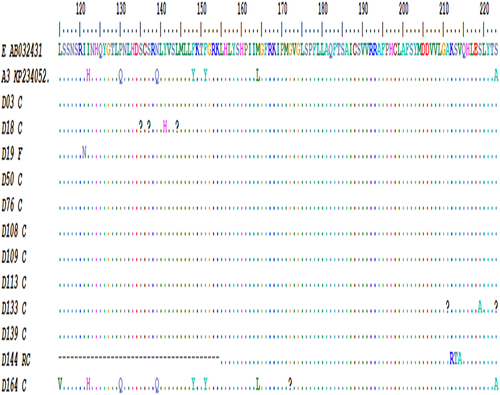
Figure 2. Rectangular phylogenetic tree depicts the rare genotype A3 sequences obtained from this study. Tree Scale: 0.05 -

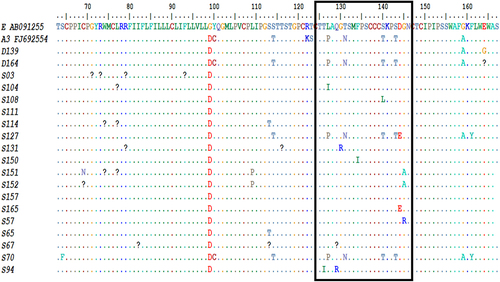
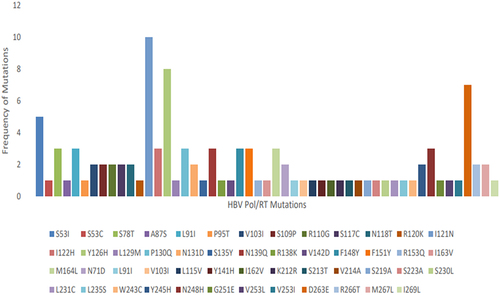
Table 2. Rate of HBV S gene pointmutation and mutation types across clinical cohorts of patients.
Table 3. Socio-demographic, serological, and HBV S gene mutational profiles of enrollees with occult HBV infection.
Supplemental Material
Download MS Word (3.9 MB)Data Availability statement
The hepatitis B virus Polymerase and S gene sequences obtained from this study had been deposited in the Genbank sequence database under accession numbers OP420514 to OP420522, OP428653 to OP428701 https://mail.google.com/mail/u/0/#search/gb-admin%40ncbi.nlm.nih.gov/FMfcgzGqQSSScVjrPsPRrPvRSnpZmLjr.