Figures & data
Figure 1. Illustration of influenza virion components, genomic organization, and viral genes. (a) Influenza virus is an enveloped RNA virus, which has 3 envelope proteins (HA, NA, and M2) on the viral membrane, an M1-formed matrix layer, and eight vRnps. (b) Each vRNP consists of one vRNA segment wrapped with NP and associated with polymerase complex PB2/PB1/PA. (c) Each vRNA segment encodes 1–3 genes, through alternative splicing (NS2/NEP and M2) and frameshifting (such as PA-X and PB1-F2). Accessory proteins expressed through frameshifting are shown as filled dark green bars/boxes.
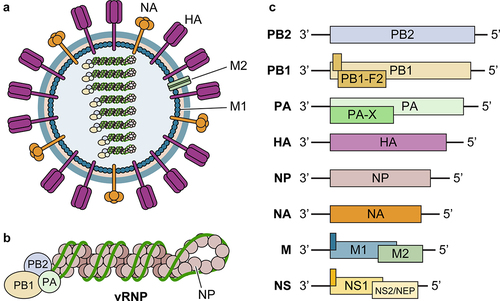
Figure 2. Influenza virus evolution. Almost all IAV subtypes (H1 to H16, N1 to N9) have natural hosts in water birds, of which some have established infections in other species, such as H1N1, H2N2, and H3N2 in humans and pigs. Human influenza viruses have changed over the years mainly due to the emergence of four pandemic flu viruses. Currently, circulating influenza viruses include two IAV subtypes H1N1 and H3N2, and two IBV lineages (B/Yamagata and B/Victoria). Some animal IAVs, particularly bird and swine flu, can occasionally spill over to cause zoonotic infections in humans. In recent years, avian H5 and H7 viruses caused human infections with a high case fatality rate.
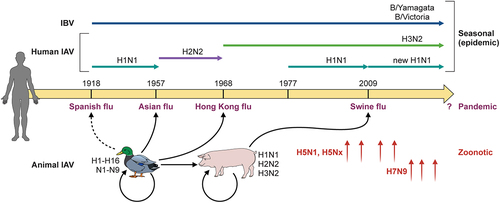
Table 1. Molecular markers associated with virulence and pathogenicity of influenza virus. HA substations are based on the H3 numbering system[Citation97].
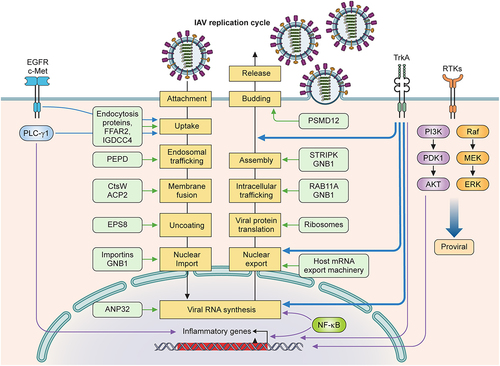
Figure 3. Pro-viral host factors and host signalling pathways important for influenza viral replication. Host factors and cellular signalling pathways are hijacked by the influenza virus to promote viral replication at different steps of the viral life cycle. Host factors are listed inside light green boxes next to the specific step of viral replication. Receptor tyrosine kinases (EGFR, c-Met, TrkA) on plasma membranes are activated by influenza viral infection and function to promote viral replication. EGFR, c-Met, and PLC-γ1 enhance viral uptake. TrkA signalling is important for several steps of the viral life cycle: viral RNA synthesis, vRNP nuclear export, and viral budding and release. NF-κB signalling enhances viral RNA synthesis and induces the expression of proinflammatory genes. PI3K/Akt and Raf/MEK/ERK pathways also strongly increase viral replication.