Figures & data
Figure 1. AFSV p17 promotes autophagy.
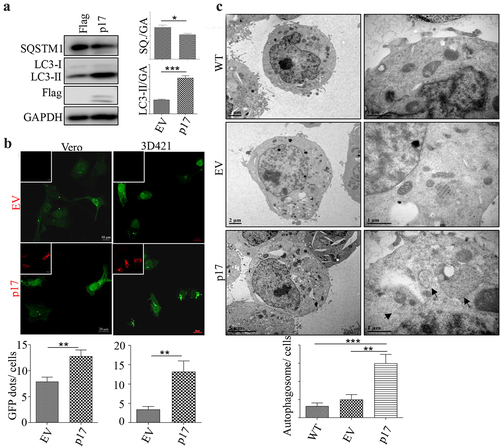
Figure 2. P17 interacts with TOMM70.
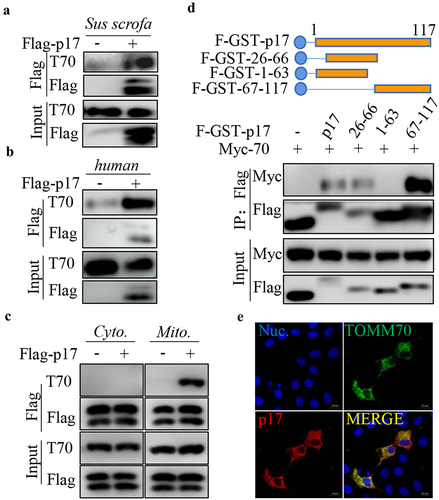
Figure 3. P17 promotes autophagic engulfment of mitochondria by facilitating the binding of SQSTM1 to TOMM70.
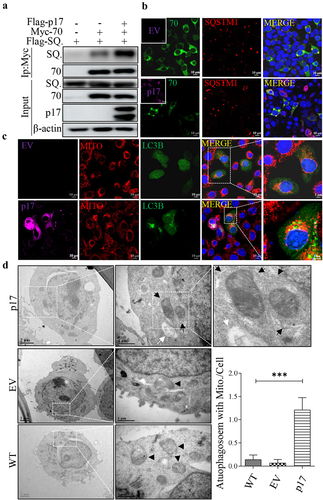
Figure 4. P17 promotes mitochondrial degradation.
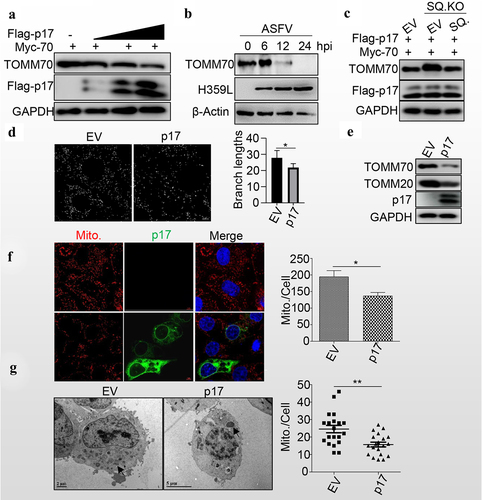
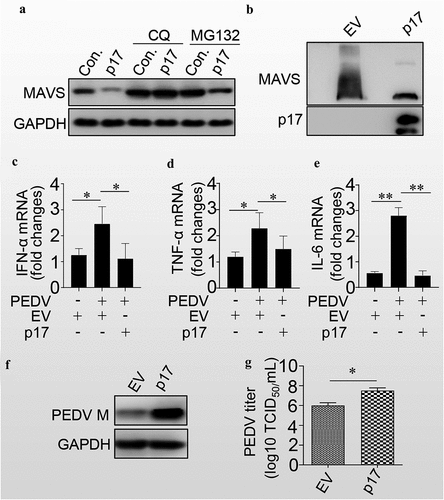
Figure 5. P17 inhibits the innate immune response by autophagic degradation of MAVS.
Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
