Figures & data
Figure 1. RopB is not involved in regulating the production of extracellular vesicles. The EVs from the overnight culture wild-type A20 strain and the ropB mutant (∆ropB) were collected by ultracentrifugation, and the particle size and number were analysed by (a) the nanoparticle tracking system (NTA) and (b) the tunable resistive pulse sensing system (qNano). (c) The morphology of EVs from A20 and ropB mutant under the scanning electron microscope observation (indicated by arrows).
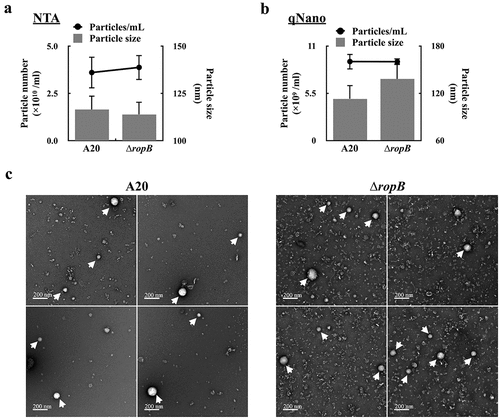
Figure 2. The SpeB cysteine protease degrades streptolysin O (SLO) in extracellular vesicles. (a) and (b) SLO in bacterial culture supernatants from the wild-type A20 strain, its ropB mutant (∆ropB), and the ropB complementary strain [∆ropB (Comp)]. (c) The expression of slo in the wild-type A20 strain and its ropB mutant. RNAs were extracted from A20 and ropB mutant after 5–16 h of incubation. The expression of slo was normalized to that of gyrA. *, P < 0.05. (d) SLO in ultracentrifuged culture supernatants (UCSs) and extracellular vesicles (EVs) from A20 and the ropB mutant. The lower panel shows the protocol for separating UCSs and EVs. (e) SLO and SpeB in EVs from A20, the ropB mutant, the speB isogenic mutant (∆speB), the SpeB protease-inactivated (SpeBC192S) mutant, and the SpeB-inducing peptide-inactivated (SIP*) mutant after 16 h of incubation. The lower panel shows the protocol for isolating EVs. Thirty-µL of supernatants and EVs were utilized for western blot hybridization. zSpeB, the zymogen form SpeB (42 kDa); mSpeB, the mature form SpeB (28 kDa). (f) The size and the number of released EVs from A20, the ropB mutant, the speB mutant, and the SpeBC192S mutant. The EVs were analysed by the nanoparticle tracking system.
![Figure 2. The SpeB cysteine protease degrades streptolysin O (SLO) in extracellular vesicles. (a) and (b) SLO in bacterial culture supernatants from the wild-type A20 strain, its ropB mutant (∆ropB), and the ropB complementary strain [∆ropB (Comp)]. (c) The expression of slo in the wild-type A20 strain and its ropB mutant. RNAs were extracted from A20 and ropB mutant after 5–16 h of incubation. The expression of slo was normalized to that of gyrA. *, P < 0.05. (d) SLO in ultracentrifuged culture supernatants (UCSs) and extracellular vesicles (EVs) from A20 and the ropB mutant. The lower panel shows the protocol for separating UCSs and EVs. (e) SLO and SpeB in EVs from A20, the ropB mutant, the speB isogenic mutant (∆speB), the SpeB protease-inactivated (SpeBC192S) mutant, and the SpeB-inducing peptide-inactivated (SIP*) mutant after 16 h of incubation. The lower panel shows the protocol for isolating EVs. Thirty-µL of supernatants and EVs were utilized for western blot hybridization. zSpeB, the zymogen form SpeB (42 kDa); mSpeB, the mature form SpeB (28 kDa). (f) The size and the number of released EVs from A20, the ropB mutant, the speB mutant, and the SpeBC192S mutant. The EVs were analysed by the nanoparticle tracking system.](/cms/asset/903cc400-8374-4727-9280-d084d7e9152a/kvir_a_2249784_f0002_oc.jpg)
Figure 3. The SpeB protease degrades bacterial proteins in extracellular vesicles. (a) Protease K protection assay for SpeB in EVs. EVs from the stationary phase A20 (after 8 h of incubation) were collected by ultracentrifugation and incubated with protease K in the presence with or without 1% SDS. SpeB was detected by western blot (the upper panel) and the lower panel shows the total protein of EVs. (b) SpeB and total protein profiles in A20’s and the speB mutant’s (∆speB) EVs after 0–24 h incubation at 37ºC. SpeB in EVs was detected by western blot (the upper panel) and the total protein profile in EVs (the middle panel) was visualized. The band density in the dash-line indicated region (the middle panel) was calculated, and the band density relative to 0 h was shown in the lower panels. zSpeB, the zymogen form SpeB (42 kDa); mSpeB, the mature form SpeB (28 kDa). *, P < 0.05. (c) The protein abundance in EVs from the speB mutant relative to that in EVs from A20. The relative protein abundance was calculated as [PSMs(∆speB) of target protein/total PSMs(∆speB)]/[PSMs of target protein(A20)/total PSMs(A20)]. The proteins with a relative abundance greater than 1.5-fold were shown. The accession number (UniProt) and the coding gene of target proteins were indicated on the x-axis. Results from the two-independent experiments were included for analysis.
![Figure 3. The SpeB protease degrades bacterial proteins in extracellular vesicles. (a) Protease K protection assay for SpeB in EVs. EVs from the stationary phase A20 (after 8 h of incubation) were collected by ultracentrifugation and incubated with protease K in the presence with or without 1% SDS. SpeB was detected by western blot (the upper panel) and the lower panel shows the total protein of EVs. (b) SpeB and total protein profiles in A20’s and the speB mutant’s (∆speB) EVs after 0–24 h incubation at 37ºC. SpeB in EVs was detected by western blot (the upper panel) and the total protein profile in EVs (the middle panel) was visualized. The band density in the dash-line indicated region (the middle panel) was calculated, and the band density relative to 0 h was shown in the lower panels. zSpeB, the zymogen form SpeB (42 kDa); mSpeB, the mature form SpeB (28 kDa). *, P < 0.05. (c) The protein abundance in EVs from the speB mutant relative to that in EVs from A20. The relative protein abundance was calculated as [PSMs(∆speB) of target protein/total PSMs(∆speB)]/[PSMs of target protein(A20)/total PSMs(A20)]. The proteins with a relative abundance greater than 1.5-fold were shown. The accession number (UniProt) and the coding gene of target proteins were indicated on the x-axis. Results from the two-independent experiments were included for analysis.](/cms/asset/376cc3e3-59ee-4e3e-8710-6b25bbbe19a1/kvir_a_2249784_f0003_b.gif)
Figure 4. Extracellular vesicle-associated SpeB fails to trigger HaCaT keratinocyte pyroptosis. (a) The cytotoxicity of the concentrated supernatant (by 10 kDa centrifugal filter) from A20 and the speB mutant (∆speB) to HaCaT cells. HaCaT cells were incubated with the 2-fold concentrated supernatants at 37ºC for 4 h. Z-vad-fmk (20 µM), the pan-caspase inhibitor. The morphology of concentrated supernatant-treated cells was shown in (b). The bubbling morphology, the typical characteristic of pyroptotic cells, is indicated by arrows. (c) Cytotoxicity of EVs from A20 and the speB mutant to HaCaT cells. HaCaT cells were treated by EVs with the multiplicity of infection (M.O.I.) 10–1000 at 37ºC for 4 h. (d) The expression of SpeB and SLO in A20, the speB mutant, and the slo mutant (∆slo). Bacterial strains were cultured to the stationary phase and the bacterial culture supernatant was analysed by western blot hybridization. zSpeB, the zymogen form SpeB (42 kDa); mSpeB, the mature form SpeB (28 kDa). (e) The cytotoxicity of the concentrated supernatant (by 10 kDa centrifugal filter) from A20, the speB mutant, and the slo mutant to HaCaT cells. The morphology of concentrated supernatant-treated cells was shown in (f) and the bubbling cell morphology was indicated by the arrow. Cytotoxicity was determined by the lactate dehydrogenase (LDH) assay. *, P < 0.05.
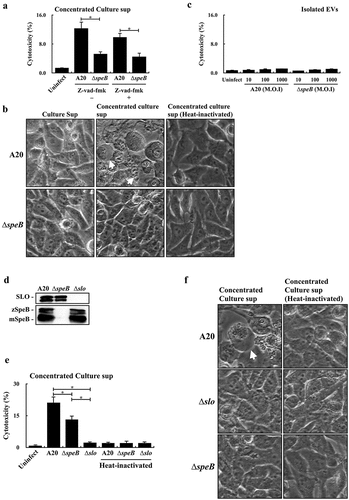
Figure 5. Enolase is resistant to SpeB degradation and could be utilized as the internal control protein in analysing GAS EVs. (a) Protease K protection assay for the unknown 45 kDa protein (indicated by the arrow) in EVs from A20 and speB mutant (∆speB). EVs from A20 and speB mutant were collected by ultracentrifugation and incubated with protease K in the presence with or without 1% SDS for 60 min at 37ºC. (b) Detection of the unknown 45 kDa protein by anti-human enolase antibody. EVs from A20 were analysed by western blot with an anti-human enolase antibody. The lower panel shows the total protein in EVs. The arrow indicates the unknown 45 kDa protein. 2×, two-fold dilution. (c) Protease K protection assay for enolase in EVs. EVs were incubated with protease K in the presence with or without 1% SDS. GAS enolase was detected by an anti-human enolase antibody and the lower panel shows the total protein of EVs. (d) Detection of enolase in EV-depleted ultracentrifuged culture supernatants (UCS). The thirty- and 15-µL UCSs and EVs from A20 were analysed by western blot with the anti-human enolase antibody. (e) Utilization of enolase as the internal control protein for analysing SLO and SpeB in EVs from A20, the speB mutant, the SpeB protease-inactivated mutant (SpeBC192S), and the slo mutant (∆slo). Thirty-µL of EVs was analysed by western blot with anti-SLO, anti-SpeB, and anti-human enolase antibodies. The lower panel shows the total protein in EVs. zSpeB, the zymogen form SpeB (42 kDa); mSpeB, the mature form SpeB (28 kDa).
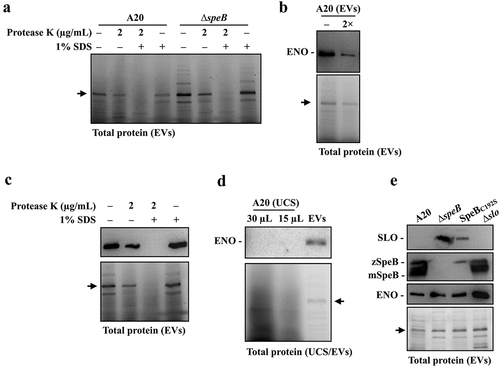
Table 1. Bacterial strains and plasmids used in this study.
Supplemental Material
Download MS Word (338.1 KB)Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article. Additional data are available from the corresponding author upon reasonable request.
