Figures & data
Figure 1. Neighbor end joining tree of L protein CDS for selected paramyxoviruses. L CDS neighbor end joining homology tree made with Tamara Nei consensus sequence of 23 sequences. L CDS from MeV (NC_001498.1), GhV (HQ660129), CedPV-CG1a (NC_025351), CedPV-Geelong (KP271122), NiV-B (AY988601), NiV-M (NC_002728), HeV (AF017149), HeV (MZ318101), AngV (ON613535), DARV (MZ574408), MojV, (NC_025352), LayV (OM101125), GAKV (MZ574409), DenV (OK623355), and MeliV (OK623354).
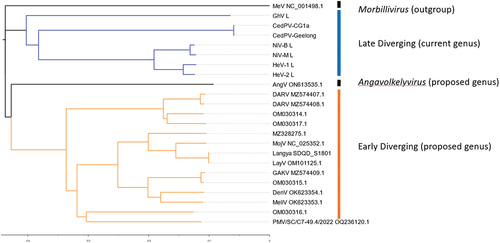
Figure 2. Genomic organization of henipaviruses and related species: of the selected genomes (a) the HeV genome has the conserved gene order N-P-M-F-G-L where each ORF is encoded on a separate mRNA which differs from (b) which is the MeliV genome characteristic of LayV, MojV, GAKV, DARV, and related viruses where the F gene encodes an additional ORF X in the 5’ UTR producing the S protein. AngV (c) has the smallest genome and each mRNA is likely monocistronic. MeV (d) which is representative of the morbillivirus genus as an outgroup is markedly smaller compared to (a) and (b). The transcriptional unit length (all nucleotides from last intergenic residue to the first residue of the next intergenic region) from each major branch of the henipavirus phylogeny compared to each other for (e) the N gene of genomes with intergenic region present in the leader, (f) for P gene in all genomes, (g) M in all genomes, (h) F transcriptional unit (including ORF X/S protein CDS for early diverging genomes), (i) G gene, (j) for the L gene in all genomes with intergenic sequence in the trailer, and (k) total reported genome length. The length of early diverging henipavirus genomes (including LayV and MojV) is longer compared to later diverging genomes (including HeV, NiV, and CedV) and the transcriptional unit encoding the F protein and/or S protein is significantly longer (f).
Table 1. Annotated genomes related to HeV and NiV. list of nearly or completely sequenced genomes related to HeV or NiV used in this review and their corresponding GenBank accession numbers.
Table 2. Current virus isolates. Currently HeV, NiV, and CedV are the only viruses for which there is a live virus isolate and reverse genetics system available. GAKV and LayV have been isolated and shown to replicate in human cells [Citation3,Citation12].
Figure 3. Map of where henipavirus related genomes have been sequenced or detected. Countries are subdivided into colours by proposed genotype, unchategorized genotypes are grey, early diverging genotypes are in Orange, late diverging genotypes are in blue. (created with mapchart.net).
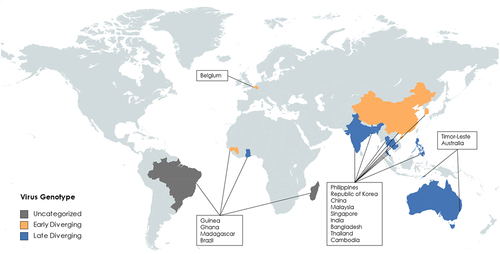
Figure 4. Comparisons of P genes from selected genotypes (a) MeV, (b) AngV, (c) LayV, MojV, GAKV, (d) NiV, HeV, GhV.
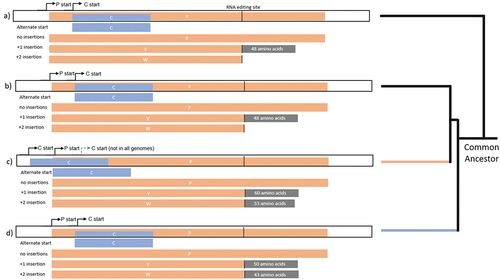
Figure 5. Predicted structures of S proteins. highest scoring Alphafold predictions with regions in blue: N-terminal and predicted intracellular region, Grey: predicted transmembrane region, Orange: predicted ectodomain of S protein CDS from (a) LayV SDQD_H1801 [OM101125], (b) Jingmen Crocidura shantungensis henipavirus 2 isolate SYS_SheQu [OM030315], (c) DewiV strain BE/Ninove/Cr/1/2019 [OK623354], (d) Melian virus strain GN/Meliandou/Cg/1/2018 [OK623353], (e) GAKV Cs17-65 [MZ574409], (f) Wufeng Crocidura attenuata henipavirus 1 isolate WFS_SheQu [OM030317], (g) MojV isolate Tongguan1 [NC_025352], (h) DARV Cl17-46 [MZ574408], (i) GAKV Cs17-65 [MZ574409], (j) MAG: chodsigoa hypsibia henipavirus isolate PMV/SC/C7-49.4/2022 [OQ236120], (k) Wenzhou apodemus agrarius henipavirus 1 [MZ328275], (l) Wufeng Chodsigoa smithii henipavirus 1 isolate WFS_ChangWei [OM030316], (m) schematic of LayV S protein [OM101125] with furin cleavage site in orange.
![Figure 5. Predicted structures of S proteins. highest scoring Alphafold predictions with regions in blue: N-terminal and predicted intracellular region, Grey: predicted transmembrane region, Orange: predicted ectodomain of S protein CDS from (a) LayV SDQD_H1801 [OM101125], (b) Jingmen Crocidura shantungensis henipavirus 2 isolate SYS_SheQu [OM030315], (c) DewiV strain BE/Ninove/Cr/1/2019 [OK623354], (d) Melian virus strain GN/Meliandou/Cg/1/2018 [OK623353], (e) GAKV Cs17-65 [MZ574409], (f) Wufeng Crocidura attenuata henipavirus 1 isolate WFS_SheQu [OM030317], (g) MojV isolate Tongguan1 [NC_025352], (h) DARV Cl17-46 [MZ574408], (i) GAKV Cs17-65 [MZ574409], (j) MAG: chodsigoa hypsibia henipavirus isolate PMV/SC/C7-49.4/2022 [OQ236120], (k) Wenzhou apodemus agrarius henipavirus 1 [MZ328275], (l) Wufeng Chodsigoa smithii henipavirus 1 isolate WFS_ChangWei [OM030316], (m) schematic of LayV S protein [OM101125] with furin cleavage site in orange.](/cms/asset/76ed7967-ca63-4b57-bfc4-7fcb2459bbbd/kvir_a_2273684_f0005_oc.jpg)
Table 3. Pathogenic differences between henipavirus outbreaks. symptoms, signs, and laboratory findings associated with outbreaks caused by Nipah virus Malaysia strain, Hendra virus genotype 1, Langya virus H1801, and an outbreak of unknown aetiology that has been speculated to be associated with Mojiang virus Tongguan1.
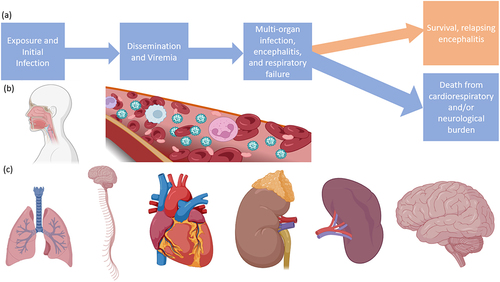
Figure 6. Progression of infection with NiV or HeV. (a) Flow chart of a typical infection, (b) routes of virus entry and dissemination, (c) organ targets including the lungs, CNS, heart, kidneys, spleen, and brain. Made with BioRender.
Data Availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
