Figures & data
Table 1. Characteristics of bacterial strains and plasmids used in this study.
Table 2. Primers used in this study. The punctuations of the primers below are incorrect. Please refer to Table 2 in our manuscript.
Figure 1. General features of S. suis PepO protease.(a) protein domains of PepO identified by SMART. (b) sequence alignments of three characteristic motifs of S. suis PepO with M. tuberculosis Zmp1, human NEP, and ECE-1. Spirals and arrows indicate helices and β-strands, respectively. Residues for substrate/inhibitor interaction are labeled with a black circle. Residues for metal coordination are labeled with a white circle. Catalytic Glu513 is labeled with a black star. (c) 3D model of PepO. Cartoon showing the α-helix, β-sheet, and random coil in lightblue, yellow, and grey. The VNAYY motif is depicted in green. Except for catalytic Glu513, the HEISH motif is depicted in red. The EIMAD motif is depicted in blue. Structure homology modelling was conducted by SWISS-MODEL using the 3D structure of Zmp1 (PDB:3ZUK) as a template.
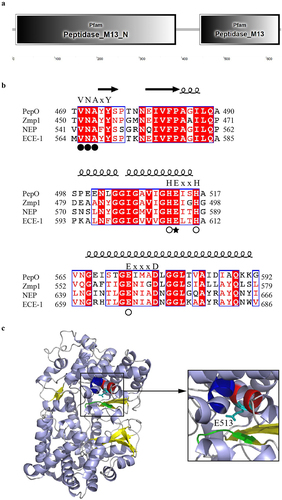
Figure 2. S. suis PepO protease degrades LL-37 and mCRAMP into shorter peptides. (a) proteolytic activity of PepO with MCA-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2 as a substrate. The Vmax and Km values were determined from the Michaelis-Menten equation. (b and c) Representative SDS-PAGE images of the proteolytic products of LL-37 (b) and mCRAMP (c) after cleavage by catalytically active PepO or inactive mutant PepOE513A. LL-37 and mCRAMP were respectively incubated with the purified PepO or PepOE513A for 3 h, the proteolytic products were analyzed using SDS-PAGE. (d-g) the MALDI-TOF/TOF mass spectrometry analyses of LL-37 (d), mCRAMP (f), the proteolytic products of LL-37 (e) and mCRAMP (g) after cleavage by the PepO. (h and i) amino acid sequences of the truncated peptides of LL-37 (h) and mCRAMP (i) after cleavage by PepO.
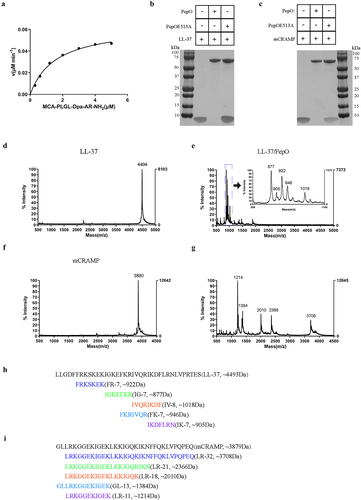
Figure 3. PepO protease protects S. suis from the bactericidal activity of LL-37 and mCRAMP. (a and b) sensitivity of S. suis strains toward LL-37 or mCRAMP. S. suis WT, ΔpepO, and CΔpepO strains were incubated with increasing concentrations of LL-37 (a) or mCRAMP (b) for 1 h. (c and d) bactericidal activities of LL-37, the proteolytic products LL-37/PepO (c), and the truncated peptides of LL-37 (d). (e and f) bactericidal activities of mCRAMP, the proteolytic products mCRAMP/PepO (e), and the truncated peptides of mCRAMP (f). The S. suis ΔpepO were exposed to different peptides or proteolytic products for 1 h. Each sample was plated on TSA to identify viable bacteria. Values represent the mean ± SD from three independent experiments. Statistical significance was calculated using two-way ANOVA tests followed by Tukey’s multiple-comparison test for panels a and b. Panels c and e were analysed via one-way ANOVA test followed by Tukey’s multiple-comparison test. Panels d and f were analysed via one-way ANOVA test followed by Dunnett’s multiple-comparison test. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
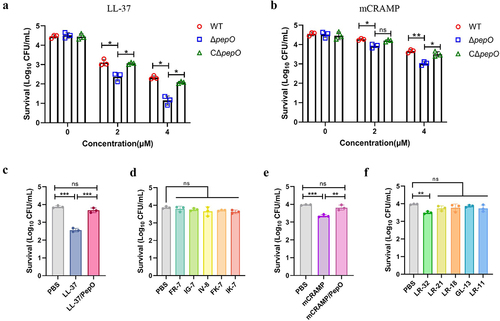
Figure 4. Cleavage of cathelicidins by PepO reduces their membrane permeabilizing activity towards S. suis. (a and b) membrane permeabilizing activities of LL-37, the proteolytic products LL-37/PepO (a), and the truncated peptides of LL-37 (b). (c and d) membrane permeabilizing activities of mCRAMP, the proteolytic products mCRAMP/PepO (c), and the truncated peptides of mCRAMP (d). The S. suis ΔpepO loaded with SYTOX green were exposed to different peptides or proteolytic products for 1 h, and the fluorescence intensity was measured. (e) microscopic visualization of the viability of S. suis ΔpepO after treatment with LL-37, mCRAMP, their proteolytic products, and their truncated peptides. Bacterial cells with intact membranes are represented by green, whereas those with damaged membranes are represented by red. Scale bars = 10 μm. Values represent the mean ± SD from three independent experiments. Statistical significance was calculated using one-way ANOVA test followed by Tukey’s multiple-comparison test for panels a and c. Panels b and d were analysed via one-way ANOVA test followed by Dunnett’s multiple-comparison test. ns, not significant; ***P < 0.001.
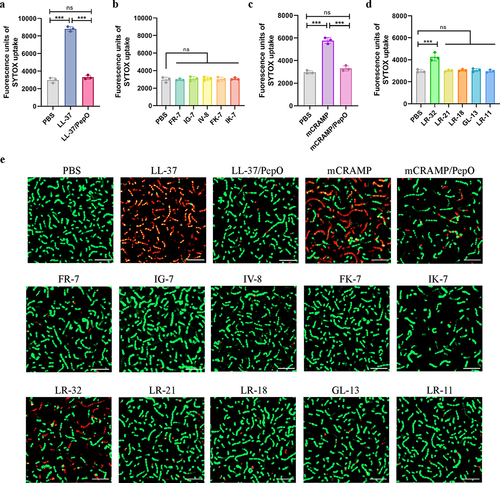
Figure 5. Cleavage of cathelicidins by PepO affects their chemotactic ability and anti-apoptotic activity towards neutrophils. (a and b) CI of neutrophils in response to LL-37, the proteolytic products LL-37/PepO (a), and the truncated peptides of LL-37 (b). (c and d) CI of neutrophils in response to mCRAMP, the proteolytic products mCRAMP/PepO (c), and the truncated peptides of mCRAMP (d). Mouse neutrophils were seeded in the upper chamber of a 5-μm Transwell, and migration was assessed after stimulation with different peptides or proteolytic products in the lower chamber. As a positive control, 5 μM of fMLP was used. The neutrophils chemotaxis toward medium was designated as 1 to calculate the CI of the different samples. (e and f) anti-apoptotic activities of LL-37, the proteolytic products LL-37/PepO (e), and the truncated peptides of LL-37 (f) toward neutrophils. (g and h) anti-apoptotic activities of mCRAMP, the proteolytic products mCRAMP/PepO (g), and the truncated peptides of mCRAMP (h) toward neutrophils. Flow cytometry was used to measure apoptosis in mouse neutrophils stimulated by TNF-α in the presence or absence of various peptides or proteolytic products. Neutrophil apoptosis in the presence of TNF-α alone was designated as 100%. Values represent the mean ± SD from three independent experiments. Statistical significance was calculated using two-tailed, unpaired t tests for panels a, c, e, and g. Panels b, d, f, and h were analyzed via one-way ANOVA test followed by Dunnett’s multiple-comparison test. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
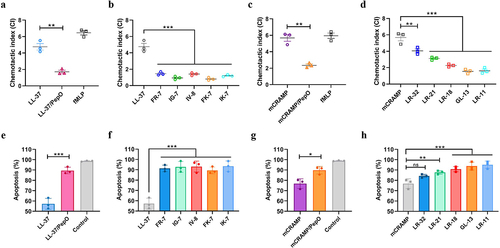
Figure 6. Cleavage of cathelicidins by PepO impairs their ability to promote lysosome formation. (a and b) microscopic visualization of lysosome formation induced by LL-37, mCRAMP, and their proteolytic products after cleavage by PepO (a), or the truncated peptides of LL-37 and mCRAMP (b). Mouse peritoneal macrophages were treated with different peptides or proteolytic products for 4 h, and stained for LAMP1 in red, nuclei in blue. Scale bars = 5 μm. (c and d) the acidic degree of lysosomes in mouse peritoneal macrophages treated with LL-37 and the proteolytic products LL-37/PepO (c), or mCRAMP and the proteolytic products mCRAMP/PepO (d) was tested by quantifying the fluorescent intensities of LysoTracker Red. Values represent the mean ± SD from three independent experiments. Statistical significance was calculated using one-way ANOVA test followed by Tukey’s multiple-comparison test. ns, not significant; *P < 0.05; **P < 0.01.
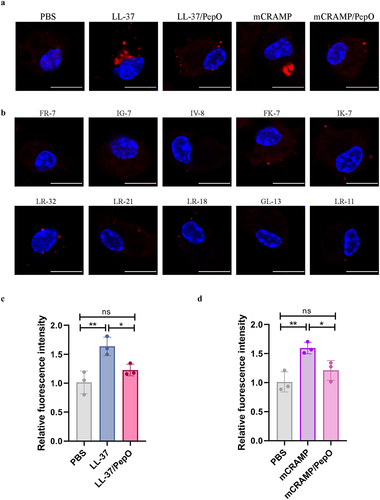
Figure 7. Loss of PepO attenuates organ injury in S. suis -infected mice. (a-d) C57BL/6 mice were inoculated with the S. suis WT strain or ΔpepO mutant by intravenous injection, and subsequently treated with or without LL-37. (a) histopathological H&E staining of brain, liver, and lung tissue sections from each group of mice. Scale bars = 100 μm. The pathological scores of the brain (b), liver (c), and lung (d) were blindly evaluated in three random fields by two independent scientists. Statistical significance was calculated using one-way ANOVA test followed by Tukey’s multiple-comparison test. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
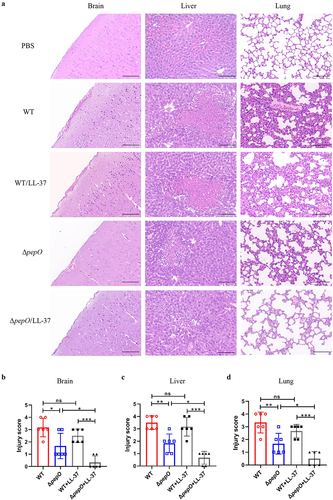
Figure 8. PepO contributes to S. suis survival and dissemination in mice. (a-d) C57BL/6 mice were inoculated with the S. suis WT strain or ΔpepO mutant by intravenous injection, and subsequently treated with or without LL-37. After 24 h of treatment, samples were collected from blood (a), lung (b), liver (c), and brain (d) for bacterial quantification. Values represent the mean ± SD from five biological replicates contained in each experimental group. Statistical significance was calculated using one-way ANOVA test followed by Tukey’s multiple-comparison test. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
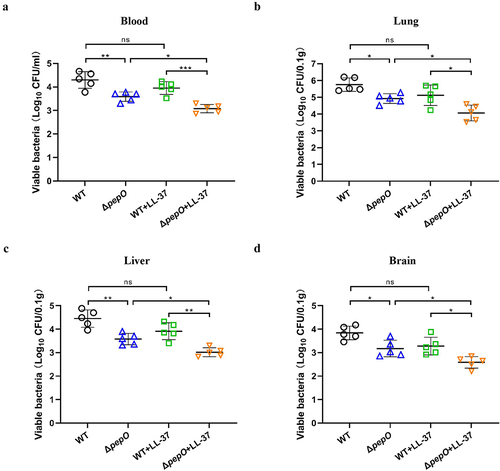
Figure 9. Schematic representation of the role of PepO-mediated cleavage of cathelicidins in S. suis immune evasion and dissemination. At the site of infection, S. suis stimulates cathelicidin production, which is stored in neutrophil granules. The mature peptide of cathelicidin can be released during neutrophil degranulation or the formation of neutrophil extracellular traps (NETs) to combat S. suis. To escape from cathelicidin-mediated antibacterial immune responses, S. suis produces an extracellular protease PepO to degrade cathelicidins LL-37 or mCRAMP into shorter fragments, abolishing its bactericidal activity against S. suis, and undermining its immunomodulatory functions, including the promotion of neutrophil migration, anti-apoptotic activity to prolong neutrophil lifespan, and the promotion of lysosome formation in macrophages. The cleavage of cathelicidins mediated by PepO promotes S. suis dissemination and exacerbates organ injury in the mouse bacteraemia model.
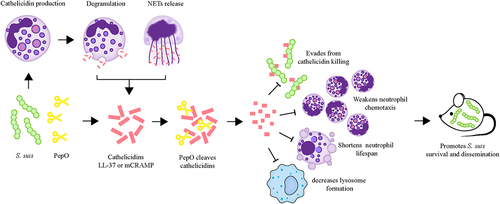
Figure S1.jpg
Download JPEG Image (104.7 KB)Data Availability statement
All the data supporting the findings of this study are provided in the article and its Supplementary file. Additional data are available upon request from the corresponding authors.
