Figures & data
Figure 1. Treponema pallidum adhered to the platelets. (a)The co-localization of platelets and T. pallidum was detected using flow cytometry. (b-d)T. pallidum-platelet attachment events were monitored using dark-field (b), scanning electron (c), and confocal immunofluorescence microscopy (d).
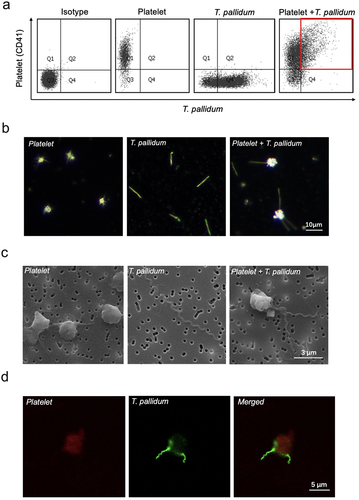
Figure 2. T. pallidum activated platelets and promoted platelet-secreting particles. (a)Activated platelets with T. pallidum were assessed using flow cytometry. (b-e)T. pallidum-induced platelets secretion of platelet factor 4 (PF4) (b), beta-thromboglobulin (β-TG) (c), platelet-derived growth factor (PDGF) (d), and endothelial growth factor (EGF) (e) was analyzed using enzyme-linked immunosorbent assay (ELISA). These data represent differentiation experiments performed across three independent experiments containing three duplicates. Data are represented as means ± standard deviation (SD) (n = 3). One-way analysis of variance (ANOVA) was used to compare multiple groups. NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
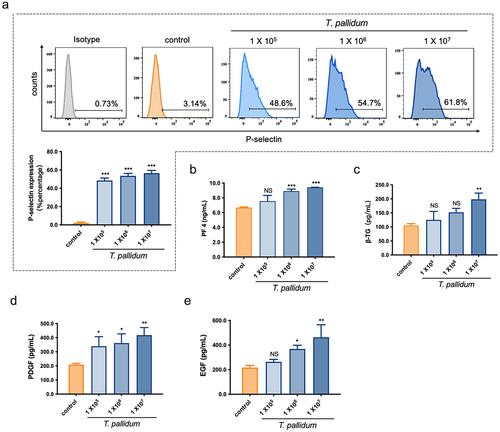
Figure 3. Pseudo-expression of major histocompatibility complex (MHC) class I on platelet-preincubated T. pallidum. (a) The secretion of MHC class I using ELISA assay. These data represent differentiation experiments performed across three independent experiments. Data are represented as means ± SD (n = 3). One-way ANOVA was used to compare multiple groups. NS, not significant; *p < 0.05. (b) The distribution of MHC class I was analyzed using confocal microscopy. (c) Analysis of MHC class I expression with post-embedding immunogold labelling (black dots) using electron microscopy.
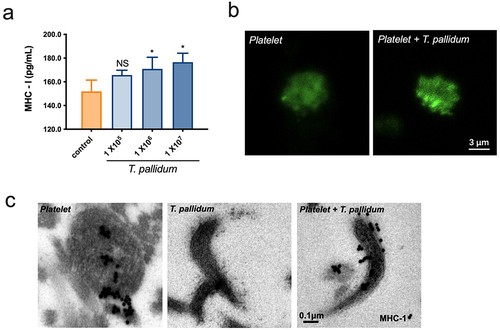
Figure 4. Preincubation of T. pallidum with platelets attenuated natural killer (NK) cell lethality. (a) Analysis of T. pallidum mRNA through targeting the polA transcript. (b) Survival rate analysis of T. pallidum. These data represent differentiation experiments performed across three independent experiments. Data are represented as means ± SD (n = 3). A student’s t-test was used to compare two groups. **p < 0.01; ***p < 0.001.
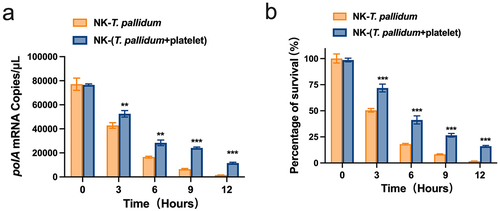
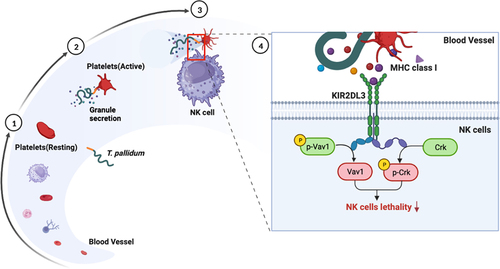
Figure 5. T. pallidum with platelets attenuated NK cell lethality via pseudo-expression of platelet-derived MHC class I. (a) Cell lysates were immunoprecipitated to analyze killer cell immunoglobulin-like receptors two immunoglobulin domains and long cytoplasmic tail 3(KIR2DL3). (b) Analysis of cell lysates for Vav1 signaling using western blotting. (c) Analysis of cell lysates for Crk signaling using western blotting. (d) Analysis of T. pallidum mRNA through targeting the polA transcript. (e) Survival rate analysis of T. pallidum. These data represent differentiation experiments performed across three independent experiments. Data are represented as means ± SD (n = 3). One-way ANOVA was used to compare multiple groups. NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
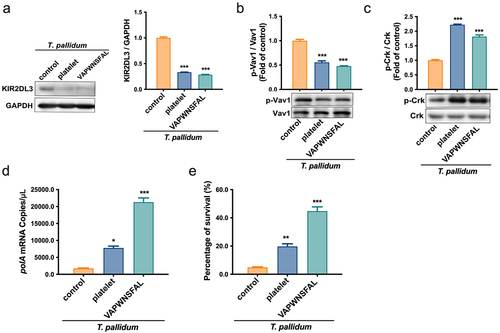
Figure 6. Schematic illustration of mechanisms underlying platelet-derived MHC class I on T. pallidum attenuated NK cell lethality. (1) T. pallidum invaded and spread through the blood. (2) T. pallidum activated platelets and promoted platelet secretion particles, while also pseudo-expressing platelet-derived MHC class I in the T. pallidum surface. (3) Pseudo-expression of platelet-derived MHC class I on T. pallidum escaped from the immune clearance of NK cells. (4) Mechanism explanation: platelet-derived MHC class I coating on T. pallidum binds to KIR2DL3 receptors of NK cells, initiates dephosphorylation of Vav1 and phosphorylation of Crk changes, and then attenuates NK cell lethality.
Data Availability statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials.
