Figures & data
Figure 1. Comparison analysis of 42 ICEHin1056 family ICEs.
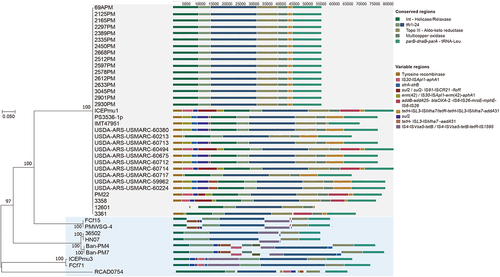
Figure 2. T4SS of 42 ICEHin1056 family ICEs in P. multocida.
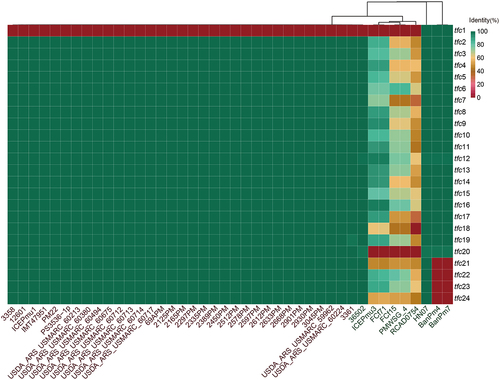
Figure 3. Collinear alignment of SXT/R39 family ICEs and Tn916 family ICEs.
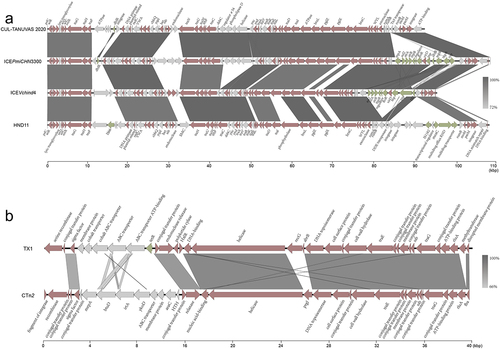
Figure 4. Module structure of resistance genes of ICEs in P. multocida.
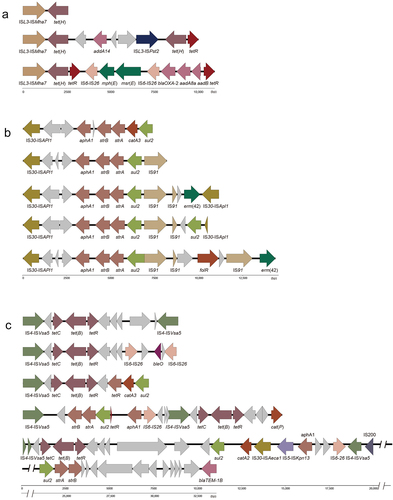
Figure 5. Prevalence analysis of ICEs in P. multocida.
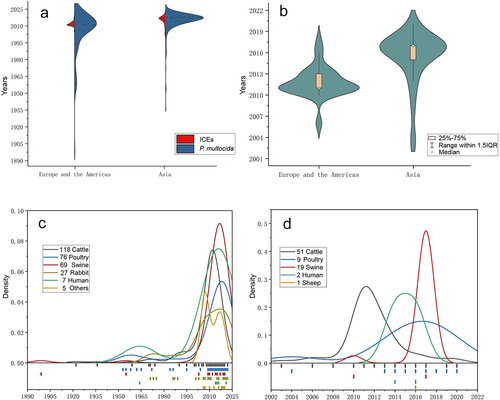
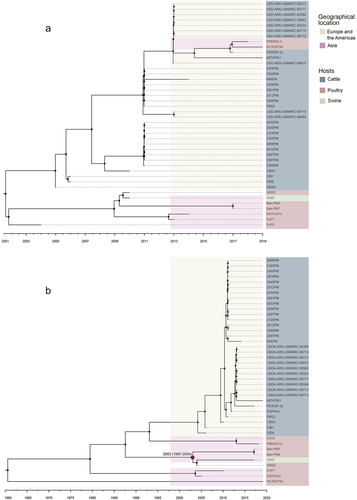
Figure 6. The Bayesian tree of ICEs and P. multocida.