Figures & data
Figure 1. Illustrative shared and distinguishing traits of Golgi important Rab proteins. Rab proteins share a GTPase cycle between nucleotide-bound, membreane associated and –free states and functional amino acid motifs colored as indicated in (A). However, activated Rab proteins recruit distinct effectors to regulate their respective pathways (B). As indicated in (C). Rab proteins have distinct locations with respect to the Golgi complex and vary in abundance as quantified for purified cell fractions.Citation8,10,13,15,17,19,21,22,28,30,39,40,41,54,56,57,58,68,76,79,86,90,93,94
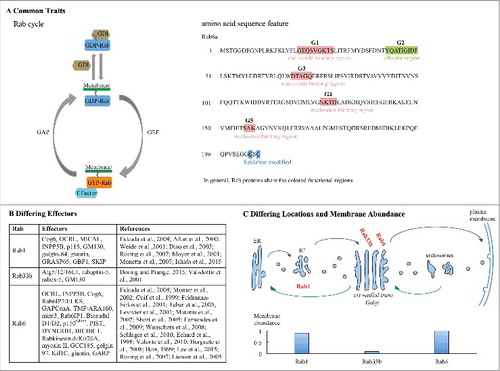
Table 1. Effect of altered rab expression on golgi ribbon organization.
