Figures & data
Figure 1. Rab22a-GFP and Rab14-RFP localization in non-polarized MDCK cells. MDCK cells were plated on glass coverslips and transfected with Rab22a-GFP (green) and Rab14-RFP (red). DAPI labels the nucleus (blue). Enlarged vesicles in the perinuclear region are both Rab22a and Rab14 positive (arrows). Scale bar, 20 μm. Lower panel is an enlargement of the inset outlined in the upper panel. Rab22a-GFP positive puncta in the cytoplasm do not contain Rab14 (arrowheads). Scale bar, 5 μm
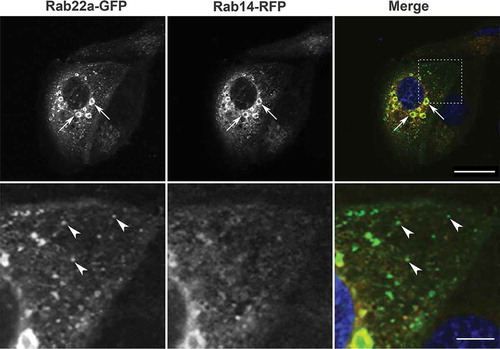
Figure 2. Rab22 localizes to the cell:cell interface in MDCK cell pairs. Cell pairs in three-dimensional culture expressing Rab22a-WT-GFP or constitutively active (Rab22a-Q64 L-GFP) together with Rab14-WT-RFP. Transfected cells were embedded in Matrigel into a chamber slide and fixed after 16 hours. Rab22a-GFP localizes to the cell:cell interface (arrows) and to puncta in the cytoplasm (arrowheads). Rab14-RFP and Rab22a-WT-GFP and Rab22a-Q64 L-GFP colocalize in the cytoplasmic puncta (arrowheads). Scale bar, 10 μm
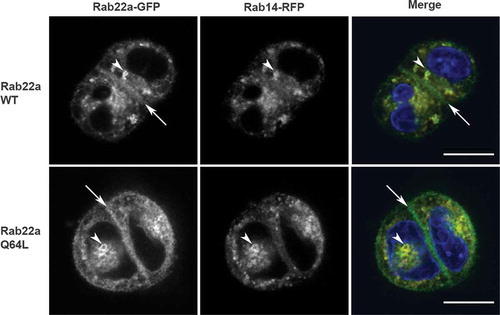
Figure 3. Knockdown of Rab22a results in multi-lumen formation and overexpression of Rab22a rescues Rab14 knockout. (a) MDCK Rab22a shRNA cells were embedded in Matrigel on a chamber slide and fixed after 48 hours. Podocalyxin (PDCX, red) was labelled to identify the forming apical domains. DAPI labels the nucleus (blue). Knockdown of Rab22a results in the formation of multiple lumens. Scale bar, 10 μm (b) Quantification of single lumen formation. The average percent of cysts with single lumen was recorded for shRNA control (n = 1050) and Rab22 knockdown (n = 817) cysts, across three independent repetitions with four replicates per plating. Rescue of single lumen with Rab22a shRNA resistant plasmid in cysts. MDCK cells that were transduced with Rab22a shRNA were transfected with Rab22a plasmid that was engineered to be resistant to the shRNA. Single cells were embedded in Matrigel and grown for 24 hours and the formation of single lumen cysts was quantified. The average percent of cysts with single lumen was recorded for shRNA control (n = 513), Rab22a knockdown (n = 606) and Rab22a shRNA with resistant plasmid (n = 190) cysts, across two independent repetitions with four replicates per plating. Expression of the Rab22a shRNA resistant plasmid rescues single lumen formation. (c) Rescue of single lumen formation in Rab14 KD cells. Rab14 KD cells were transfected with Rab22a-WT-GFP and Rab22a-Q67 L-GRP and single lumen formation was quantified. The average percent of cysts with single lumen was recorded for shRNA control (n = 304), Rab14 shRNA (n = 483), Rab14 shRNA with Rab22a-WT-GFP (n = 125), and Rab14 shRNA with Rab22a-Q64 L-GFP (n = 149) cysts across two independent repetitions with four replicates per plating
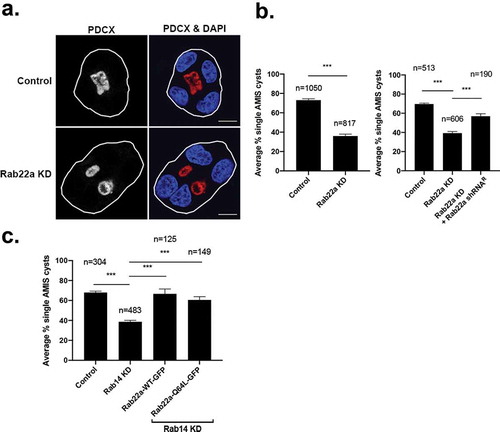
Figure 4. Rab22a does not colocalize with PDCX, but knockdown of Rab22a delays its trafficking to the apical membrane. (a) Rab22a shRNA and control cells were embedded in Matrigel. Cells were fixed after 16 hours and labelled for PDCX (red). Rab22a KD results in delayed trafficking of PDCX from the periphery to the forming lumen, DAPI labels the nucleus (blue). Scale bar, 10 μm (b) Quantification of localization of PDCX in cell pairs of Rab22a shRNA (n = 233) and control cells (n = 219). PDCX location was determined as either completely within the cell at the interface, completely on the peripheral cell membrane, or both. The percentage of cell pairs represents the average percentages across the three independent repetitions of the experiment. (c) WT cells were transfected with Rab22a-GFP and plated on glass coverslips. Cells were fixed and labelled for PDCX (red). The distribution of Rab22a-GFP and PDCX are distinct. (d) WT cells were transfected with Rab22a-Q64 L-GFP and plated on glass coverslips. Cells were fixed and labelled for PDCX (red). The distribution of Rab22a-Q64 L-GFP and PDCX are distinct. Lower panels are an enlargement of the inset outlined in the upper panel. Distinct puncta that are only Rab22a-GFP positive (arrow) or PDCX positive (arrowhead) are seen and colocalization is not observed. Average Pearson’s correlation coefficient = 0.31 for Rab22a-WT-GFP, and average Pearson’s correlation coefficient = 0.12 for Rab22a-Q64 L-GFP. Scale bar (upper panels, 5 μm, lower panels, 5 m). (e) WT MDCK cells were transfected with Rab22a-WT-GFP and plated in Matrigel. Cells were fixed after 16 hours and labelled for PDCX (red). Scale bar, 10 μm. (f) WT MDCK cells were transfected with Rab22a-Q64 L-GFP and plated in Matrigel. Cells were fixed after 16 hours and labelled for PDCX (red). Scale bar, 10 μm
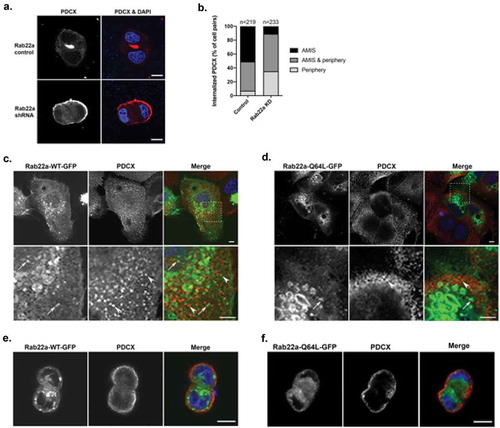
Figure 5. Rab22a knockdown results in decreased Arf6 activity and Rab22a interacts with EFA6. (a) Rab22a shRNA and control cells were lysed and subjected to pull down to assess levels of active Arf6. (b) Quantification of the active-Arf6 pull down. Pull down experiments were performed in three independent experiments. (c) Rab22a co-immunoprecipitates with EFA6 but not ARNO. HEK cells expressing EFA6-Flag and either Rab22a-WT-GFP or Rab22a-Q64L-GFP or ARNO-myc and Rab22a-Q64L-GFP were precipitated with antibodies against the Flag or myc tag and blotted for Rab22a and Flag/myc
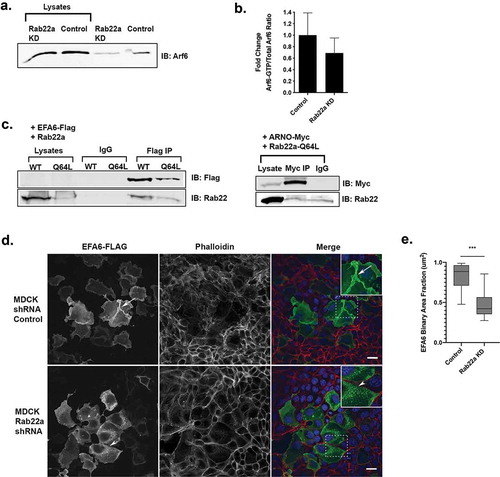
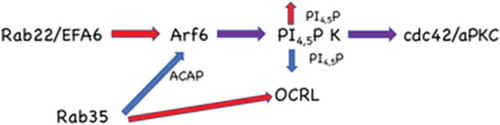
Figure 6. Model for Rab22a function in the establishment of polarity. Rab22a delivers EFA6 to the forming apical membrane, activating Arf6, resulting in increased phosphatidylinositol 4-phosphate 5-kinase activity and thus PtdIns4,5P2. Rab35 negatively regulates Arf6 activity through the Arf6 GAP ACAP2. Rab35 also regulates the phosphatidylinositol 4,5-bisphosphatase, OCRL. Both of these Rab35 regulated activities would decrease PtdIns4,5P2 at the plasma membrane. This tight regulation of the lipid composition allows the appropriate recruitment of polarity proteins such as Cdc42 and aPKC