Figures & data
Figure 1. Two mutations in ELMO3, arising from biallelic heterozygosity in a patient, abrogate the GEF activity of the ELMO3/DOCK1 complex. a) Pedigree schematics. Filled symbol = affected individual. The mutation status is mentioned under each individual: + = WT allele, M1 and M2 = mutant alleles as defined in b). b) Human ELMO3 gene schematic with nucleotide position of M1 and M2 mutations pointed with an arrow. c) Schematic representation of ELMO3 and its functional domains: Ras-Binding Domain (RBD), ELMO inhibitory domain (EID), ELMO domain (ELMO), PH domain (PH), ELMO auto-regulatory domain (EAD) and proline-rich motif (PxxP). The mutations in ELMO3 reported in this study, Val337Ile and Ser385Cys, are found close or within the ELMO domain, and are each encoded by a single mutant allele of the biallelic heterozygosity state in the patient. d, e) Functional studies performed to characterize the biochemical properties of each of the ELMO3 mutants (each experiment was performed n = 3). d) The individual ELMO3 mutants co-immunoprecipitated with DOCK1 when co-expressed in HEK293T cells. e) Active RAC1 was analysed via a GST-PAK-PBD pull-down assay. f) Quantification of the assay done in e). The expression of the ELMO3 mutants/DOCK1 complex led to less RAC1 activation in comparison to the exogenous expression of the wild-type ELMO3/DOCK1. Two-tailed unpaired t test was used; *P < 0.05.
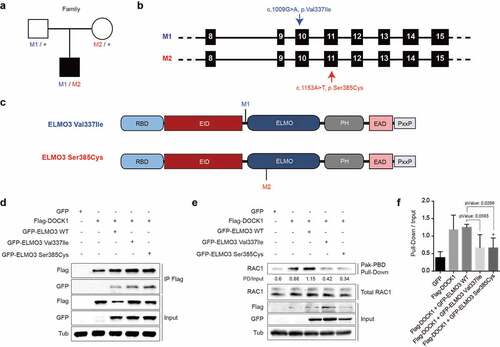
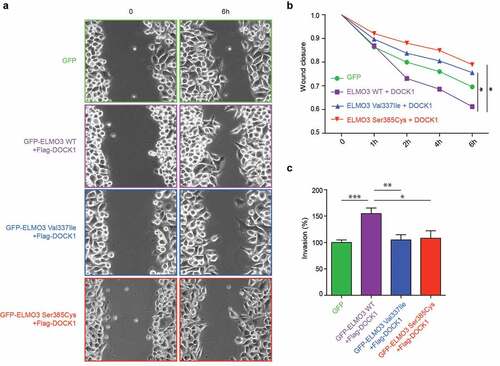
Figure 2. The individual patient-derived ELMO3 mutations impair cell migration and invasion. a) HeLa cells expressing Flag-DOCK1 with wild-type GFP-ELMO3 showed increased cell migration in comparison to cells expressing GFP alone, while Flag-DOCK1 co-expressed with either of the ELMO3 mutants showed less cell migration in a wound healing assay (n = 3, *p = 0.0319 and **p = 0.0019). b) Quantification of the wound closures shown in (a). c) Boyden chamber Matrigel invasion assays were performed with HeLa cells expressing wild-type or ELMO3 mutants together with DOCK1, or as a control, GFP alone. DOCK1/ELMO3 mutants showed an impaired ability to promote invasion in comparison to DOCK1/ELMO3 WT (n = 3, ***p < 0.0001, **p = 0.0017 and *p = 0.0134).