Figures & data
Figure 1. MDS for a KRAS template. Unique barcodes and an adaptor are introduced into, for example, exon 1 of KRAS through restriction digestion (❶) and first-strand extension (❷), followed by linear amplification to copy the original DNA 12 times (❸) and 20 cycles of exponential amplification (❹) to expand the library for sequencing (❺). Sequencing reads are grouped by barcode and bona fide mutations (●) separated from false ones (◯) by virtue of being present in all reads sharing the same barcode. Adapted from Refs. [Citation10,Citation11].
![Figure 1. MDS for a KRAS template. Unique barcodes and an adaptor are introduced into, for example, exon 1 of KRAS through restriction digestion (❶) and first-strand extension (❷), followed by linear amplification to copy the original DNA 12 times (❸) and 20 cycles of exponential amplification (❹) to expand the library for sequencing (❺). Sequencing reads are grouped by barcode and bona fide mutations (●) separated from false ones (◯) by virtue of being present in all reads sharing the same barcode. Adapted from Refs. [Citation10,Citation11].](/cms/asset/faaf177b-6d12-45f2-8bdc-99f54d1b303a/ksgt_a_2083895_f0001_oc.jpg)
Table 1. Frequency of mutations engineered into KRAS cDNA templates spiked into 293 T genomic DNA.
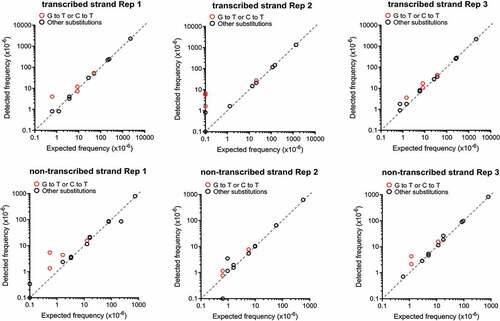
Figure 2. MDS accurately detects KRAS cDNA mutants spiked into 293T genomic DNA. Frequency of single (detected) versus co-occurring (expected) mutations identified by MDS using a dilution series of KRAS cDNAs containing two different mutations engineered in KRAS exon 1 mixed with genomic DNA from 293T cells. Three biological replicates for MDS assay performed on the transcribed strand of KRAS exon 1 are show on the top and three biological replicates for MDS assay performed on the non-transcribed strand of KRAS exon 1 are show on the bottom.