Figures & data
Figure 1. RhoA-G14V activates mDIA1 and ROCK2 but not Rhotekin. A) Schematic illustration of the constructs for producing GST-fusion effector RBDs. RTKN-RBD: RBD of mouse Rhotekin (amino acids 7–89); ROCK2-RBD: RBD of human ROCK2 (amino acids 979–1068); mDIA1-RBD: RBD of human mDIA1 (amino acids 69–451). B) Production of GST-fusion RBDs. The indicated GST fusion proteins were produced in BL21 E.coli and captured by Glutathione Sepharose beads. The proteins eluted from the beads were stained with GelCode Blue after electrophoresis on an SDS-PAGE gel. C-D) Detection of active RhoA by GST pulldown assay. HEK293 cells were transfected with HA-tagged wild-type (WT) and mutant RhoA expression vectors as indicated. Lysates from the transfected cells were extracted and subjected to GST pulldown assay using the GST-RTKN-RBD, GST-ROCK2-RBD, or GST-mDIA1-RBD beads. Both active (GTP-bound) and total RhoA were detected by anti-RhoA antibody, and ⍺-Tubulin was probed for loading controls. E) Detection of RhoA activation by GST pulldown assay using RIPA buffer [Citation3]. HEK293 cells transfected with Mock, HA-RhoA-Q63L, or HA-RhoA-G14V were lysed in the RIPA buffer, and subjected to GST pulldown assay using GST-RTKN-RBD or GST-mDia1-RBD beads. F) Assessment of RhoA-induced SRF-driven luciferase reporter activity in HEK293 cells co-transfected with SRF-Luc, Renilla, and the indicated RhoA expression vector. The assay was performed in triplicate, and the bars represent average luciferase activity. Error bars represent standard deviation (***: p < 0.0005; ns: not significant).
![Figure 1. RhoA-G14V activates mDIA1 and ROCK2 but not Rhotekin. A) Schematic illustration of the constructs for producing GST-fusion effector RBDs. RTKN-RBD: RBD of mouse Rhotekin (amino acids 7–89); ROCK2-RBD: RBD of human ROCK2 (amino acids 979–1068); mDIA1-RBD: RBD of human mDIA1 (amino acids 69–451). B) Production of GST-fusion RBDs. The indicated GST fusion proteins were produced in BL21 E.coli and captured by Glutathione Sepharose beads. The proteins eluted from the beads were stained with GelCode Blue after electrophoresis on an SDS-PAGE gel. C-D) Detection of active RhoA by GST pulldown assay. HEK293 cells were transfected with HA-tagged wild-type (WT) and mutant RhoA expression vectors as indicated. Lysates from the transfected cells were extracted and subjected to GST pulldown assay using the GST-RTKN-RBD, GST-ROCK2-RBD, or GST-mDIA1-RBD beads. Both active (GTP-bound) and total RhoA were detected by anti-RhoA antibody, and ⍺-Tubulin was probed for loading controls. E) Detection of RhoA activation by GST pulldown assay using RIPA buffer [Citation3]. HEK293 cells transfected with Mock, HA-RhoA-Q63L, or HA-RhoA-G14V were lysed in the RIPA buffer, and subjected to GST pulldown assay using GST-RTKN-RBD or GST-mDia1-RBD beads. F) Assessment of RhoA-induced SRF-driven luciferase reporter activity in HEK293 cells co-transfected with SRF-Luc, Renilla, and the indicated RhoA expression vector. The assay was performed in triplicate, and the bars represent average luciferase activity. Error bars represent standard deviation (***: p < 0.0005; ns: not significant).](/cms/asset/fab43946-5e03-4932-84ce-53f2eae7557c/ksgt_a_2111945_f0001_oc.jpg)
Figure 2. RhoA downstream effectors are differentially activated by different stimuli. A) Representative Western blots showing detection of LPA- and FBS-induced RhoA activation by three different effector-based GST pulldown assays. HEK293 or Hela cells were stimulated with LPA (1 μM) or FBS (5%) for 5 min and 10 min after serum-starvation, and the cell lysates were subjected to GST pulldown assay described above. B) Quantification of RhoA activation derived from three independent data sets. Box bars represent fold changes of the ratio of active RhoA/total RhoA by comparing stimulated vs control. Error bars represent standard error. Statistical significance was determined by student t-test (*: p < 0.05; **: p < 0.005; ***: p < 0.0005). C) Representative Western blots showing detection of RhoA activation upon colchicine treatment. GST pulldown assays were performed with the lysates of HEK293 and Hela cells treated with colchicine (10 μg/ml) for 5’, 10’, 30’, and 60’. As controls, cells were treated with DMSO (the solvent for colchicine). D) Quantification of three independent RhoA activation assays upon colchicine treatment.
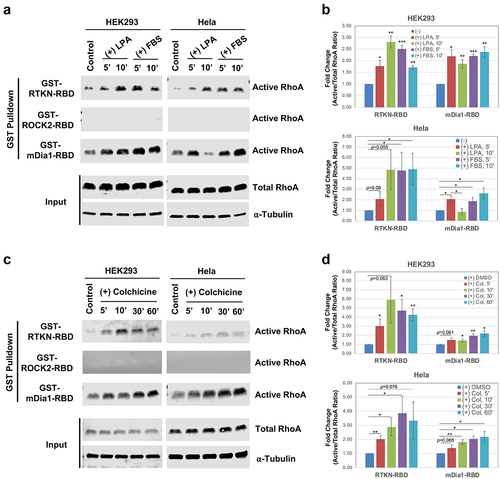
Figure 3. BRET assay development. A) Diagram depicting the principle of the BRET assay. B) Construct design for BRET assay donors and acceptors. Donors were constructed by fusion of RhoA to the N-terminus or C-terminus of Nluc, and the acceptors were constructed by fusion of effector RBD to the N-terminus or C-terminus of mVenus. All fusion constructs were linked by an in-frame flexible peptide linker (GGGGSGGGGS). C-D) Western blot detection of the indicated donor and acceptor proteins. Nluc and its fusion proteins were detected by anti-Luc antibody, and the mVenus and its fusion products were detected by anti-GFP antibody. Loading controls were probed with anti-⍺ Tubulin. E) Detection of RhoA activation by BRET assay in HEK293 cells after co-transfection of the indicated N-terminally linked Nluc-fused RhoA (Nluc.L-RhoA, WT and mutants) with the N-terminally linked Venus-fused effector RBDs (Venus.L-RBDs). Statistical significance was determined by student t-test, comparing the corresponding RhoA co-transfected with Venus-fused RTKN-RBD or mDIA1-RBD to those co-transfected with the backbone Venus (*: p < 0.05; **: p < 0.005; ***: p < 0.0005). F) BRET assay for the C-terminally linked Nluc-fused wild type RhoA (RhoA-WT-L-Nluc) co-transfected with the indicated Venus-fused RBD vectors (both N-terminally and C-terminally linked). G) Titration of Venus.L-RTKN-RBD (0, 1 ng, 5 ng, 25 ng, and 100 ng) for detection of BRET activity after co-transfection with the Nluc.L-RhoA (WT and mutants) (1 ng) in HEK293 cells. The amount of Venus vector used for each transfection was same (total 100 ng) after adjusted with backbone Venus accordingly.
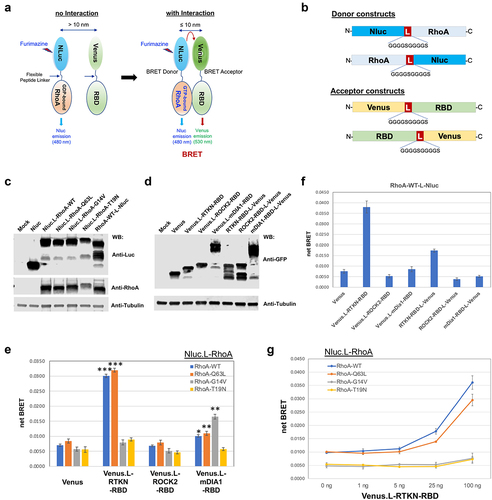
Figure 4. Assessment of stimulus-induced RhoA activation by BRET assay. A) HEK293 cells were co-transfected with Nluc.L-RhoA-WT and the indicated Venus fusion vectors. BRET activity was measured at 5 min and 10 min after cells were stimulated with LPA (1 μM), FBS (5%), or colchicine (10 μg/ml). B) BRET assay for HEK293 cells with the indicated four different combination of transfection with/without LPA (1 μM) stimulation for 5 min.
Supplemental Material
Download Zip (1 MB)Data availability statement
No data were deposited in a public database. Request for reagents used in this study should be addressed to J.E. Casanova ([email protected])
