Figures & data
Figure 1. Wild-type Gtr2p is incapable of crosslinking to nucleotides. (a) A Coomassie stained gel showing the purified Gtr heterodimers from S. cerevisiae and S. pombe. (b) Crosslinking assays probing the binding of GTP (left panels) or GDP (right panels) to wild-type Gtr heterodimers from S. cerevisiae and S. pombe. Gtr1p is capable of being resolved in all cases, but not Gtr2p. (c) Quantification of GTP (circle) and GDP (square) binding to S. cerevisiae Gtr1p, fit to a single-site binding equation. (d) Quantification of GTP (circle) and GDP (square) binding to S. pombe Gtr1p, fit to a single-site binding equation. (e) Crosslinking assays probing the binding of GTP to GTP-binding deficient Gtr mutants, Gtr1p(S20N)-Gtr2p and Gtr1p-Gtr2p(S23N). When Gtr1p is the only subunit capable of binding GTP, we can resolve the crosslinked bands. When Gtr2p is the only subunit capable of binding GTP, no obvious crosslinked bands are observed.
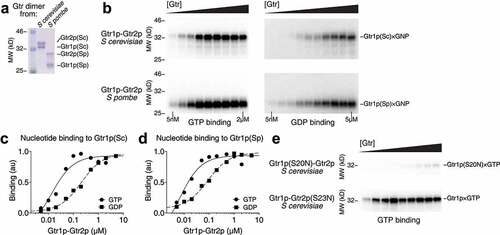
Table 1. Summary of nucleotide-binding constants (Kd) at 4°C.
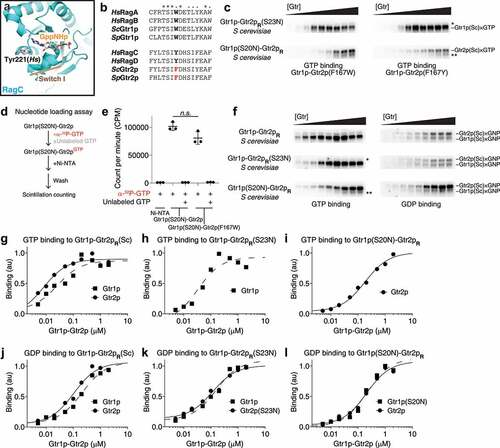
Figure 2. A mutation on Gtr2p restores the nucleotide crosslinking capabilities. (a) Crystal structure of the nucleotide-binding domain of human RagC (3LLU) reveals a tyrosine residue (Tyr221) in close proximity to the bound nucleotides. (b) Sequence conservation analysis of the Tyr221 residue on human RagC. A conserved aromatic residue occupies this position in Rag/Gtr GTPases. (c) GTP-binding assays using Gtr heterodimers carrying F167W (left panels) or F167Y (right panels) mutation on Gtr2p. The crosslinked bands corresponding to Gtr2p bound to GTP can be clearly resolved here. (d) Reaction scheme for the GTP loading experiment. (e) Quantification of the amounts of radioactively labelled GTP that remain bound to the GTPase by scintillation counting. Ni-NTA column is the blank control without any Gtr1p-Gtr2p. Columns 2 and 4: radioactively labelled GTP loading onto Gtr1p(S20N)-Gtr2p and Gtr1p(S20N)-Gtr2p(F167W) heterodimer, respectively. No significant change was observed. Columns 3 and 5: radioactively labelled GTP loading onto Gtr1p(S20N)-Gtr2p and Gtr1p(S20N)-Gtr2p(F167W) heterodimer in the presence of unlabelled GTP as a competitor. No radioactively labelled GTP was detected, suggesting the specificity of the binding event. (f) Nucleotide binding to Gtr1p-Gtr2pR. S. cerevisiae wild-type, Gtr1p(S20N)-Gtr2p, and Gtr1p-Gtr2p(S23N) heterodimers carrying F167W mutation were used. (g-l) Quantifications of the nucleotide-binding assays shown in panel D. The data were fit to a single-site binding equation.