Figures & data
Figure 1. CD8+BTLA+ TIL exhibit a less differentiated, more activated phenotype than their CD8+BTLA− counterparts. (A) Enrichment analysis of a global set of genes from CD8+ BLTA+ or BTLA− subsets. The top 50 genes that are immunologically relevant in either the BTLA+ or BTLA− subset are shown, ranked by enrichment scores using gene-set enrichment analysis (GSEA). Each row represents 1 gene and each column represents 1 sample of sorted CD8+ BTLA+ or BTLA− subset from 1 patient. TIL were isolated from melanoma metastases, cultured with 3,000 IU/mL IL-2 for 3 wks and stained for expression of CD8, BTLA, CD45RA, CCR7, CD28, CD27, PD-1, TIM3, granzyme B and perforin. Dead cells were excluded using Aqua® live/dead dye. (B, left panel) CD45RA and CCR7 expression on the CD8+BTLA+ (left dot plot) and CD8+BTLA− (right dot plot) populations on a representative TIL from patient 2360. (B, right panel) A summary of CD45RA and CCR7 expressions within each CD8+BTLA+ or CD8+BTLA− subset (n=11). Differentiation subsets are defined as follows: TN: CD45RA+CCR7+; TCM: CD45RA+CCR7+, TEM: CD45RA−CCR7−, TEMRA: CD45RA+CCR7−. The results are the percentages of TEM or TEMRA subsets within each gated CD8+ BTLA+ or BTLA− subset. TN and TCM are omitted because no significant populations were found. (C) Summary of the expression of surface markers CD28, PD-1, and TIM3 by the CD8+BTLA+ and CD8+BTLA− subsets (n ranges from 6–42 per marker). Statistical significance between the subsets was determined using Wilcoxon signed-rank test.
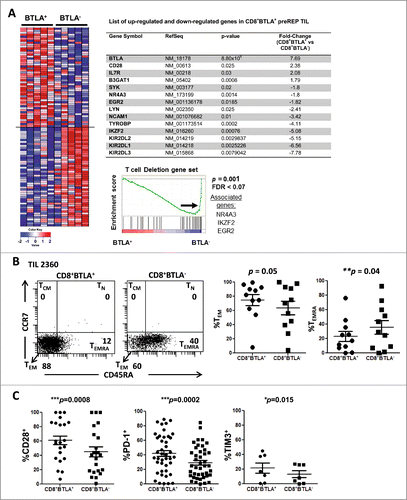
Figure 2. CD8+BTLA+ TIL have enhanced proliferation in response to IL-2 stimulation compared to the CD8+BTLA− subset. Sorted CD8+BTLA+ and CD8+BTLA− TIL subsets were labeled with carboxyfluorescein succinimidyl ester (CFSE) and cultured at a density of 1 × 106/mL with high-dose IL-2 for 5 d (3,000 IU/mL) (n = 4). (A) Histograms showing percentages of proliferating cells from 2 lines (top). The absolute number of cells in each subset was determined using trypan blue exclusion and graphed as fold expansion (bottom). * indicates significance (P < 0.05) as determined by the Student t test.
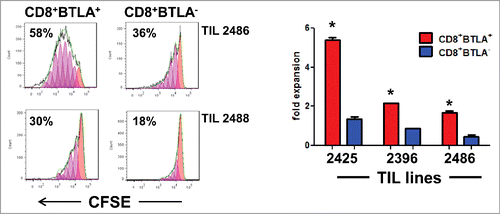
Figure 3. Enhanced IL-2 responsiveness by CD8+BTLA+ TIL. (A) TIL were stained for expression of CD8, BTLA, and CD25 and with the Aqua® live/dead viability dye. The left dot plot shows representative staining of TIL 2538 and 2547. The numbers indicated percent expression within the AQUA−CD8+ population. The right graphs summarize the percentage CD25 expression and CD25 mean fluorescence intensity (MFI) for the CD8+BTLA+ and CD8+BTLA− subsets (n = 23). P value was calculated using Wilcoxon signed-rank test. (B) TIL were sorted into CD8+BTLA+ and CD8+BTLA− subsets and cultured with increasing concentrations of IL-2 for 20 min. Cells were fixed, permeabilized, and stained for pStat5 (Y694). The left histograms show the level of pSTAT5 (Y694) expression after treatment with 200 IU/mL IL-2 for 20 min. The change in MFI (ΔMFI) between the subsets is shown. The top graphs show the change in percent pSTAT5 levels after incubation with increasing concentrations of IL-2 for 3 independent TIL lines. The shift in pSTAT5 MFI was determined by subtracting the baseline levels for each subset (n = 4). Statistical significance was determined using a paired Student t test.
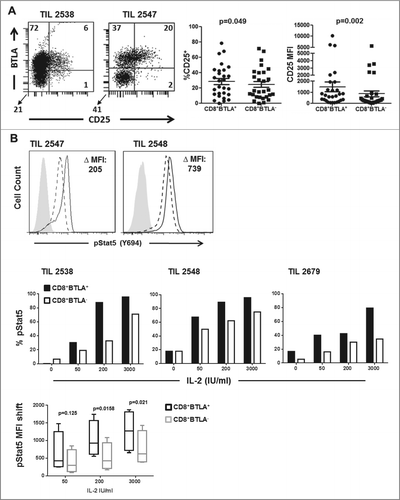
Figure 4. CD8+BTLA+ TIL are more responsive to TCR stimulation and produce more IL-2. (A) Sorted CD8+BTLA+ and CD8+BTLA− cells were stimulated with plate-bound anti-CD3 (OKT3) and anti-CD28 (OKT3+CD28) or not stimulated (NIL) in the absence of IL-2 for 3 d Cells were pulsed with 1 μCi of [3H]-thymidine for the last 18 h of culture. Results were shown as counts per minute (cpm) from triplicate wells (mean ± SD). * indicates significance (P < 0.05) as determined by Student t test. (B) Sorted CD8+ BTLA+ and CD8+BTLA− subsets were stimulated with 200 IU/mL IL-2, OKT3, or OKT3 plus CD28 for 72 h. Cells were analyzed for expression of CD25, LIGHT, and PD-1 by flow cytometry. The percent expression of each marker is shown. (C) Sorted CD8+ BTLA+ and CD8+BTLA− subsets were stimulated with PMA and ionomycin for 4 h and stained for expression of intracellular IL-2. The numbers indicate the MFI for each subset from 2 TIL lines.
![Figure 4. CD8+BTLA+ TIL are more responsive to TCR stimulation and produce more IL-2. (A) Sorted CD8+BTLA+ and CD8+BTLA− cells were stimulated with plate-bound anti-CD3 (OKT3) and anti-CD28 (OKT3+CD28) or not stimulated (NIL) in the absence of IL-2 for 3 d Cells were pulsed with 1 μCi of [3H]-thymidine for the last 18 h of culture. Results were shown as counts per minute (cpm) from triplicate wells (mean ± SD). * indicates significance (P < 0.05) as determined by Student t test. (B) Sorted CD8+ BTLA+ and CD8+BTLA− subsets were stimulated with 200 IU/mL IL-2, OKT3, or OKT3 plus CD28 for 72 h. Cells were analyzed for expression of CD25, LIGHT, and PD-1 by flow cytometry. The percent expression of each marker is shown. (C) Sorted CD8+ BTLA+ and CD8+BTLA− subsets were stimulated with PMA and ionomycin for 4 h and stained for expression of intracellular IL-2. The numbers indicate the MFI for each subset from 2 TIL lines.](/cms/asset/ac78ba86-40e9-4f5a-959e-6e3c867dc817/koni_a_1014246_f0004_b.gif)
Figure 5. In vivo tracking of infused CD8+BTLA+ and CD8+BTLA− subsets. TIL (100 × 106 cells) from the infusion product of treated patients were sorted into bulk CD8+ and CD8+ BTLA+ and CD8+BTLA− subsets and flash frozen. PBMCs collected at the labeled time points were treated in the same manner. DNA was isolated from the banked samples as described in Methods and sent for CDR3 sequencing at Adaptive Biotechnologies. (A) Vβ TCR diversity shown as pie charts for each subset with the total number of Vβ clonotypes found. (B) Overlap of each CDR3 sequence for the sorted CD8+BTLA+ and CD8+BTLA− subsets from the infusion product. Overlapping CDR3s are shown in blue, whereas CDR3 sequences found only in the CD8+BTLA+ subset are shown in red, and those found only in the CD8+BTLA− subset are shown in green. The axis shows the frequency of each CDR3 sequence for each subset within the infusion product. The percentage of the infusion product consisting of overlapping subsets is indicated. (C) Frequency of the unique CD8+BTLA+ and CD8+BTLA− subsets present in the blood over time.
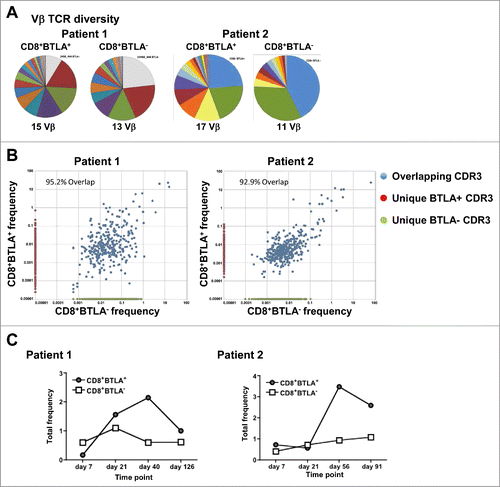
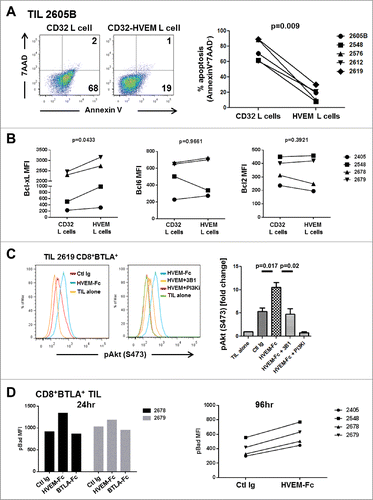
Figure 6. For figure legend, see page 10.Figure 6 (see previous page). BTLA ligation provides a prosurvival signal to TIL. (A) CD32+ L cells or CD32+HVEM+ L cells (HVEM L cells) were pulsed with 200 ng/mL OKT3 and cultured with TIL (1×106/mL) in the presence of 100 IU/mL IL-2. After 5 days, TIL were harvested and stained for CD8, Annexin V, and 7-AAD. The left dot plots show representative staining of a TIL line gated on CD8+ cells. The right graph shows the percentage of apoptosis among all lines tested. Statistical significance was determined using a Wilcoxon signed-ranked test. (B) Sorted CD8+ BTLA+ TIL were cultured with L cells as in (A) and stained for Bcl-xL, Bcl-6, and Bcl-2 after 5 d (n = 4). (C) TIL were sorted into CD8+BTLA+ and CD8+BTLA− subsets and stimulated for 2 h with 30 ng/mL OKT3 and 10 μg/mL HVEM-Fc (plate-bound) (HVEM-Fc+OKT3), 10 μg/mL anti-BTLA blocking antibody, 3B1 (HVEM-Fc+OKT3+3B1), and 10 nM PI3K inhibitor (GSK2126458) (HVEM-Fc+OKT3+PI3Ki). TIL cultured alone or with OKT3 and a control Ig were included as negative controls. TIL were immediately fixed with prewarmed Phosflow Fix buffer I for 10 min at 37°C, permeabilized with pre-chilled Phosflow Perm buffer III, and stained with pAkt (S473) for 30 min on ice. The left histograms show representative staining from TIL 2619. The right graphs show the fold change in percent pAkt expression normalized to the TIL-alone condition (set to 1; n = 3 TIL lines). Statistical significance was determined using a 2-tailed paired Student t test. (D) CD8+ BTLA+ TIL were cultured with 30 ng/mL OKT3 plus control Ig (Ctl Ig), OKT3 plus 10 μg/mL HVEM-Fc (HVEM-Fc), or OKT3 plus 10 μg/mL BTLA-Fc (BTLA-Fc) for 24 and 96 h. Cells were stained for the presence of pBad by intracellular flow cytometry as described in Methods.