Figures & data
Figure 1. Combining IL-15 with IL-12 enhanced the cytolytic activity and IFNγ production capacity of HPC-NK cells toward AML. The functionalities of HPC-NK cell products were assessed after overnight co-culture with target cells. (A) Percentage killing of K562, THP-1, and KG-1a cells at increasing effector cell:target (E:T) ratio. Combined results (mean ± SEM) of 7 independent experiments, each performed in triplicate with different HPC-NK cell donors, are shown. (B-D) Granzyme B, IFNγ, and TNFα levels were determined by ELISA in co-culture supernatants of 5 × 104 NK cells with target cells at a E:T ratio of 1:1. Mean values ± SEM of 5 donors are shown. (E) Percentage killing (mean ± SD) of patient-derived primary AML cells (E:T = 3:1). The cytolytic activity of HPC-NK cells derived from donor I (undetermined HLA-C type) was addressed using a CFSE-based cytotoxicity assay, whereas viable AML cells were identified by flow cytometry using 7AAD and CD33/CD34/CD56 markers for donor II (HLA-C group C1/C2). Statistical analysis was performed using a 2-way ANOVA (with repeated measurements in panels A-D), *P < 0.05, **P < 0.01, ***P < 0.001.
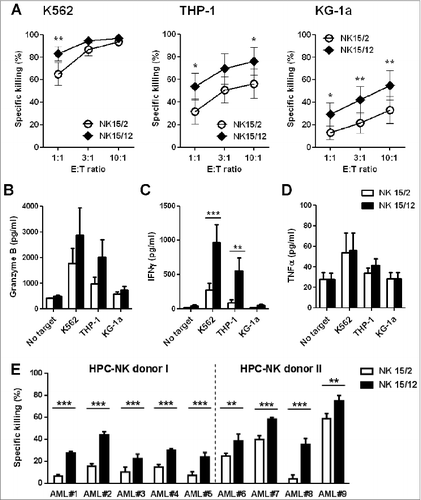
Figure 2 (See previous page). Replacement of IL-2 by IL-12 favors the development of more mature and highly functional KIR+ natural killer cells. (A) Representative expression level of TRAIL and FasL on CD56+ HPC-NK cells. Numbers indicate ΔMFI between isotype control and specific staining. (B) Relative gene expression of the cytolytic mediators perforin (PRF1) and granzyme B (GrzB), and the transcription factors eomesodermin (EOMES) and t-bet (TBX21). Mean values ± SD of 2 HPC-NK cell donors are shown, one generated with GBGM and the second one using SCGM. (C-D) Percentages of degranulating and IFNγ-producing NK cells were determined upon overnight stimulation with K562 cells at an E:T ratio of 1:1 and in the presence of low dose IL-15 (5 ng/mL). Percentages of CD107+ and IFNγ+ cells were determined with subtraction of signals measured with unstimulated cells. One representative experiment (C) and a summary of 10 donors (D, mean ± SEM) are shown. (E-F) The IFNγ production capacity of NK15/12 cells was examined within different subsets identified using NKG2A and KIRs differentiation markers. One representative donor (E) and a summary of 4 donors (F, mean ± SEM) are shown. Differences were analyzed using a paired t-test (D) or one-way ANOVA (F), *P < 0.05, **P < 0.01.
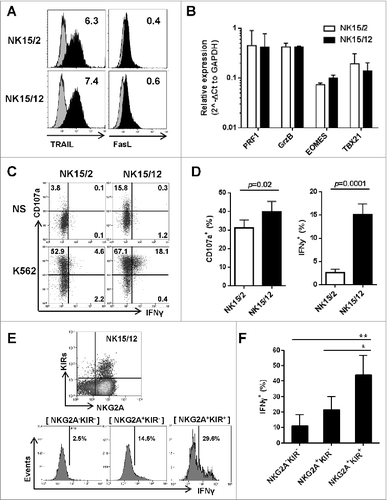
Figure 3. IFNγ-mediated upregulation of ICAM-1 on AML cells strengthens NK cell cytolytic activity. (A) Expression of ICAM-1 and LFA-3 on AML cell lines after 24 h culture with increasing concentrations of IFNγ and TNFα. (B) Expression profile of CD11a on NK15/2 and NK15/12 cells at the end of the culture process. Histograms from one representative HPC-NK cell donor out of 3 analyzed are shown. (C) Overnight THP-1 and KG-1a cell killing by NK15/2 versus NK15/12 cells, or NK15/2 cells in the presence of 1,000 pg/mL recombinant IFNγ (E:T ratio 3:1, mean ± SD). (D) Inhibition of HPC-NK cell killing in the presence of anti-ICAM-1 antibody (10 µg/mL). (E-G) Co-culture experiments of NK15/2 or NK15/12 cells with patient-derived primary AML cells performed at an E:T ratio 2:1 and in the presence of low dose IL-15 (5 ng/mL) for 24, 48, and 72 h. (E) Relative ICAM-1 expression level on AML cells and (F) relative killing (mean ± SD) of AML cells overtime upon co-culture with NK cells. The NK specific killing of AML cells was determined at each time point based on the number of surviving AML cells alone as described in the Materials and Methods section, and was depicted as relative killing compared to NK15/2 cells. (G) Expression of CD11a on HPC-NK cells upon co-culture with AML cells measured at 48 h. Data shown in (E-G) were obtained with AML#7 cells. Statistical analysis was performed using a one-way ANOVA (C) and Student t test (D, F), **P < 0.01, ***P < 0.001, ****P < 0.0001.
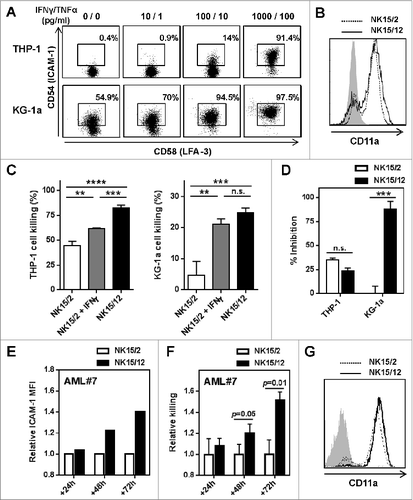
Figure 4. HPC-NK cell products generated in the presence of IL-12 display faster maturation in vivo. NK15/2 or NK15/12 cells were infused into adult NSG mice (5 × 10Citation6) in combination with low-dose IL-15 support. One week later, mice were sacrificed and ex vivo flow cytometric analysis was performed on cells isolated from spleen, liver, and bone marrow (BM). (A) Representative expression of the activating receptors NKG2D, NKp46, and DNAM-1 as well as CXCR3 on NK15/2 and NK15/12 cells before infusion and isolated from spleen 1 week after adoptive transfer. (B) Percentages of CD16- and KIR-expressing NK cells before infusion and 1 week after adoptive transfer. Combined results from 2 experiments (n = 4–6 mice per group per experiment) are shown as means ± SEM. **P < 0.01, 2-way ANOVA. (C) Representative dot plots of CD16 and KIR expression on CD56+ cells isolated from spleen of mice injected with NK15/2 or NK15/12 cells.
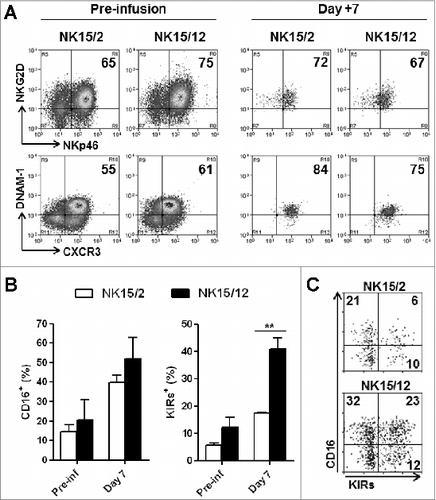
Figure 5. NK15/12 cells undergo concomitant CD16 and KIR acquisition in vivo with sustained NKG2A expression. HPC-NK cells (5 × 106) were infused into adult NSG mice in combination with IL-15 support. Ex vivo flow cytometric analysis was performed on cells isolated 1, 7, and 14 d after adoptive transfer (n = 4 per time point). (A) Percentages of splenic NK cell subsets identified using CD16 and KIR differentiation markers over time. **P < 0.01, ***P < 0.001, one-way ANOVA. (B) Percentages of liver CD16+KIR+ cells following infusion of NK15/2 versus NK15/12 cell products. *P < 0.05, ***P < 0.001, 2-way ANOVA. (C) Percentages (mean ± SD) of NK cell subsets identified using NKG2A and KIR differentiation markers over time. (D) Representative dot plots of KIR, NKG2A, and CD16 expression on human CD56+ cells of the infused product and splenocytes isolated at day 1 and 14.
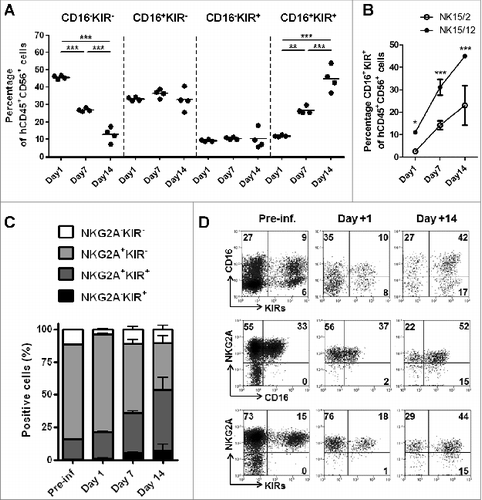
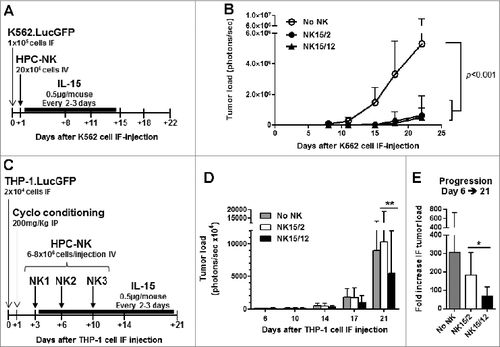
Figure 6. Infusion of NK15/12 cell products improves anti-leukemic response in THP1-bearing NSG mice. (A, B) In vivo antileukemic potential of a single infusion of NK15/2 versus NK15/12 cells was compared in adult NSG mice injected in the right femur with K562.LucGFP cells. (A) Experimental study design in K562-bearing mice, and (B) tumor load over time (mean ± SD, n=6 per group, 2-way ANOVA). (C-E) Effect of multiple NK cell infusions in cyclophosphamide-conditioned mice. Data from one experiment in which HPC-NK cells were generated using SCGM and selected for partial KIR/ligand mismatch (UCB HLA type: Bw4, C1/C2, THP-1 HLA type: Bw6, C1/C1) are shown (n=8–9 per group). (C) Experimental study design. (D) Tumor load progression overview. Mean ± SD, 2-way ANOVA with repeated measures. (E) Fold increase in tumor load between day 6 and day 21. Mean ± SD, one-way ANOVA. *P < 0.05, **P < 0.01.