Figures & data
Figure 2. Heatmaps of the selected genes in the TNBC training set (upper left) and the TNBC validation sets (upper right: validation, lower left: Ignatiadis, lower right: METABRIC).
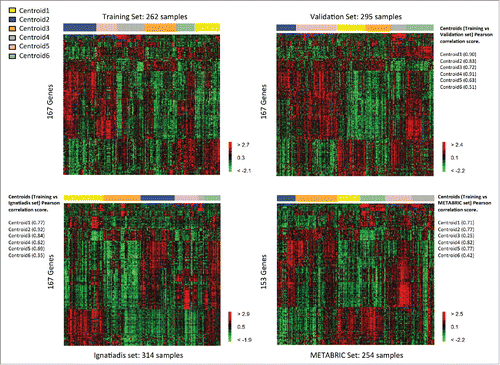
Table 1 A. Survival analysis (disease-specific survival). Chemotherapy-treated population. Univariate and multivariate analysis.
Table 1 B. Survival analysis (disease-specific survival). Chemotherapy-treated population. Two univariate models. Combination of NPI score and Immunity2 metagene expression
Figure 4. String Software connections between our Immunity1 and Immunity2 genes and the genes of eight previously published prognostic immune signatures. Stronger associations between genes are represented by thicker lines. Associations between genes with a coefficient < 0.9 are shown in green. Associations between genes with a coefficient ≥0.9 are shown in red. Associations between genes with a coefficient between 0.4 to 0.7 are not shown.
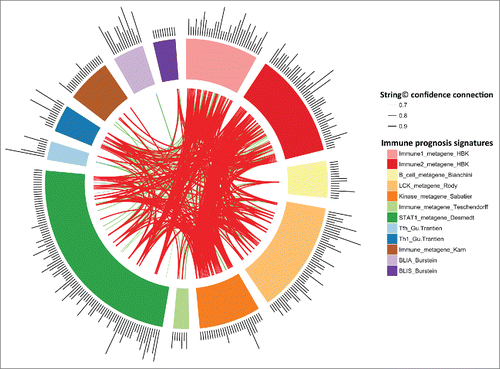
Table 2 A. Survival analysis (disease-specific survival). Chemotherapy-naive population. Univariate and multivariate analysis.
Table 2 B. Survival analysis (disease-specific survival). Chemotherapy-naive population. Univariate analysis. Combination of NPI score and Immunity2 metagene expression.
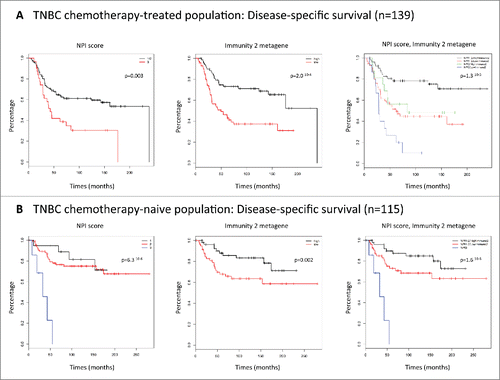
Figure 3. (A) Kaplan–Meier plots. Disease-specific survival of the chemotherapy-treated population (n = 139). NPI score. Immunity2 metagene. NPI score/Immunity2 metagene. (B) Kaplan–Meier plots. Disease-specific survival of the noCT population (n = 115). NPI score. Immunity2 metagene. NPI score/Immunity2 metagene.