Figures & data
Figure 1. T-MPs polarize macrophages toward M2 phenotype under normoxia. (A) Real-time PCR analysis of arginase1 mRNA expression in monocytes, bone marrow-derived M0, M2 and M1 macrophages, primary peritoneal macrophages and Raw264.7 cell line treated with H22-MPs for 24 h under normoxic condition. (B) The expression of CD206, IL-10 and CD301 in M0 and M2 macrophages treated with H22-MPs for 24 h under normoxic condition was analyzed by flow cytometry. (C) The expression of NOS2, TNF-α, IL12p35 and IL12p40 in M1 macrophages treated with H22-MPs for 24 h under normoxic condition was detected by real-time PCR. (D) Flow cytometry analysis the expression of TNF-α and IL-12 in M1 macrophages treated with H22-MPs. (E) The expression of iNOS in M1 macrophages treated with H22-MPs for 24 h under normoxic condition was analyzed by protein gel blot. (F) M0 macrophages were treated with T-MPs from H22 tumor cells under UV irradiation or hypoxic condition. IL-4 was used as a positive control. (G) The arginase1 expression in M0 macrophages treated with MPs generated from liver and spleen cells under normoxic and hypoxic condition. MPs generated from liver and spleen cells were added to M0 macrophages for 24 h, and then the expression of arginase1 was analyzed by real-time PCR. (H) MPs or lysate derived from H22, LLC, CT26, B16 and 5 mM lactic acid were added to M0 macrophages for 24 h, the expression of arginase1 was detected by real-time PCR. All experiments were performed at least twice. The histogram bars represent the expression level of three biological replicates, displayed as means±s.e.m. *p <0.05, ***p <0.001. NS, not statistically significant.
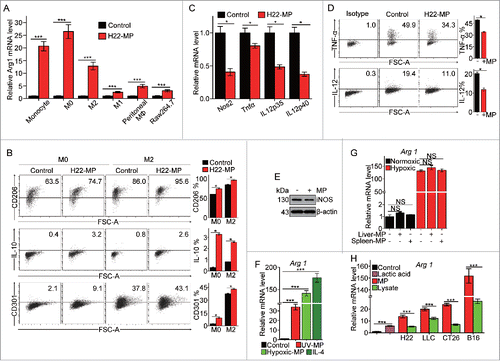
Figure 2. H22-MPs promote M2 macrophage proliferation but induce M1apoptosis. (A) 5 × 105 M0 and M2 macrophages were treated with H22-MPs and cell number was analyzed at 24 h and 48 h, respectively. (B, C) M0 and M2 macrophages were treated with H22-MPs for 24 h, and then cells were strained with FITC-Annexin V and PI for apoptosis analysis (B) or labeled with FITC-conjugated anti-BrdU antibody for cell cycle analysis (C). (D, E) 5 × 105 M1 macrophages were treated with H22-MPs and cell number was analyzed (D). Meanwhile, cells were strained with FITC-Annexin V and PtdIns for flow cytometric analysis of apoptosis (E). Data shown are representative of three reproducible experiments expressed as means±s.e.m. *p < 0.05, **p < 0.01, NS, not statistically significant.
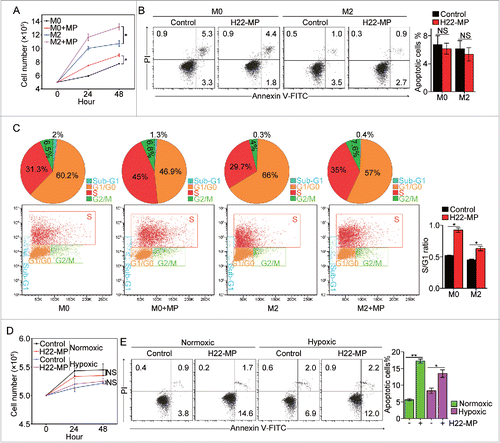
Figure 3. T-MP-educated macrophages promote tumor growth and metastasis. (A) 5 × 104 H22 tumor cells were injected to the right thigh muscle of mice. Six days later, 1×107 H22-MPs were injected to either peripheral or central site of tumors once per day for three times. On day 15, mice were sacrificed and the tumor weight was measured. (B) 5 × 104 H22 tumor cells were injected to the right thigh muscle of BALB/c mice on day 1. Clodronate (FormuMax Scientific Inc., CA) was i.p. injected into mice on day 5 (200 μL) and day 8 (100 μL), respectively. 1 × 107 H22-MPs were injected to peripheral site of tumors on day 7. On day 14, mice were sacrificed and the tumor weight was measured. (C) T-MP-educated macrophages promote tumor growth in vivo. M0, M2 and Raw264.7 macrophages were incubated with H22-MPs for 24 h, respectively. Thereafter, 2 × 105 H22 plus 6 × 104 treated or untreated macrophages were subcutaneously injected into BALB/c mice for tumor growth. The growth of tumor was monitored (n = 6, each group). (D) H22-MP-educated macrophages promote H22 tumor cell growth in vitro. M0 and M2 macrophages were incubated with H22-MPs for 24 h, and then H22 cells were added to the above group, H22 tumor cell number was analyzed at different time points. (E–G) H22-MP-educated macrophages promote melanoma metastasis. 5 × 104 B16 tumor cells plus 1 × 104 H22-MP-treated or untreated M0 macrophages were intravenously injected into C57BL/6 mice. Three weeks later, the formed tumor nodules in the lungs were examined (E), and apart from lung metastasis, metastatic tumors in various sites were analyzed (F). Meanwhile, the long-term survival of mice was analyzed by Kaplan–Meier analysis (G). (H) H22-MP-educated macrophages promote H22 tumor metastasis in BALB/c mice. 2 × 105 H22 tumor cells plus 6 × 104 H22-MP-treated M0 macrophages were intravenously injected into BALB/c mice. Metastatic tumors in various sites were analyzed. Data shown are representative of three reproducible experiments expressed as means±s.e.m. *p < 0.05, **p < 0.01, NS, not statistically significant.
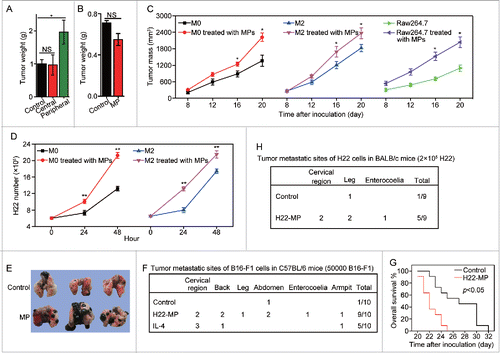
Figure 4. T-MP-educated macrophages promote TRCs growth via producing MFGE8 and TGF-β1. (A–C) H22-MPs-educated macrophages increased colony size and number of H22 TRCs. M0 macrophages were treated with or without H22-MPs for 24 h and the supernatants were used to culture H22 tumor cells in 3D fibrin gels for TRC growth. On day 5, the colony size of H22 TRCs was visualized by microscope (A) and analyzed by Image J software (B). Meanwhile, the colony number of H22 TRCs was analyzed by microscope (C). (D) The expression of Bmi1, CD44, Hif1α and c-myc in H22 TRCs was detected by real-time PCR. (E) The expression of MFGE8 and TGFB1 in H22-MPs-treated or untreated macrophages was analyzed by real-time PCR. (F–H) Downregulating the expression of MFGE8 and TGFB1 in H22-MPs-educated macrophage inhibits H22 TRCs colony size and number. The scale bar represents 100 μm. Data shown are representative of three reproducible experiments expressed as means±s.e.m. *p < 0.05, **p < 0.01, *** p < 0.001.
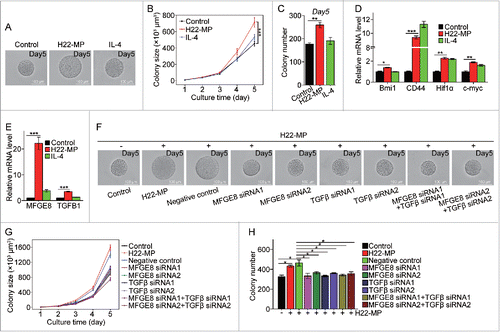
Figure 5. T-MPs polarize M2 macrophages through the cGAS/STING pathway. (A) The expression of arginase1 and phosphorylation of TBK1, STAT6 and STAT3 in M0 macrophages treated with H22-MPs were detected by Western blot. (B) The expression of STAT6 and arginase1 in M0 macrophages transfected with STAT6 siRNAs was detected by real-time PCR. (C, D) The expression of TBK1, arginase1 or STAT6 in M0 macrophages transfected with TBK1 siRNAs was analyzed by real-time PCR (C) and Western blot (D). (E) The expression of STING and arginase1 in M0 macrophages transfected with STING siRNAs was analyzed by real-time PCR. (F) The expression of cGAS and arginase1 in M0 macrophages transfected with cGAS siRNA was analyzed by real-time PCR. Data shown are representative of three independent experiments expressed as means ± s.e.m. *p < 0.05, ***p < 0.001.
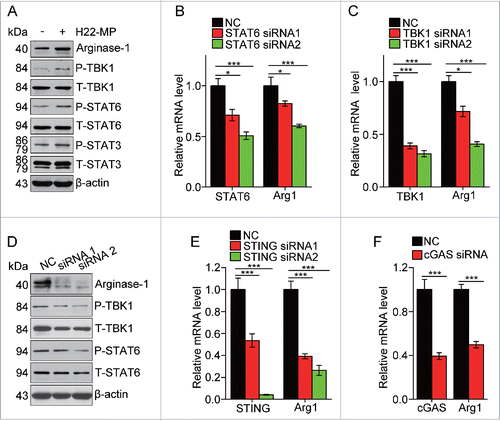
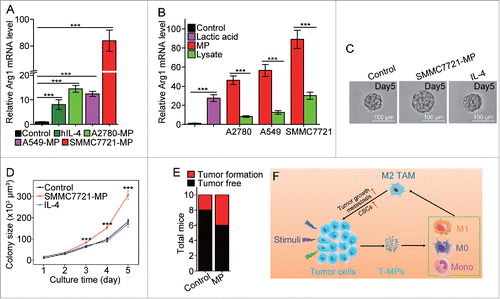
Figure 6. SMMC7721-MPs induce the differentiation of PMA-stimulated THP-1 cells toward M2 phenotype. (A) THP-1 cells treated with 100 ng/mL PMA for 48 h were incubated with hIL-4, A2780-MPs, A549-MPs, SMMC7721-MPs for 24 h and the expression of arginase1 was detected with real-time PCR. (B) The expression of arginase 1 in PMA-stimulated THP-1 cells treated with A2780-MPs, A2780-lysate, A549-MPs, A549-lysate, SMMC7721-MPs, SMMC7721-lysate and 5 mM lactic acid was analyzed with real-time PCR. (C, D) SMMC7721-MPs-educated PMA-stimulated THP-1 cells promote SMMC7721 TRCs colony size. PMA-stimulated THP-1 cells were treated with or without SMMC7721-MPs for 24 h and the supernatants were used to culture SMMC7721 tumor cells in 3D fibrin gels for TRC growth. On day 5, the colony size of SMMC7721 TRCs was visualized under microscope (C) and analyzed by Image J software (D). (E) 1 × 106 SMMC7721 tumor cells plus 3 × 105 SMMC7721-MPs-treated or untreated PMA-stimulated THP-1 cells were subcutaneously injected to nude mice (n = 10 per group). On day 35, the tumor formation was obtained. (F) T-MPs act as a general mechanism to polarize macrophages into M2 type tumor-associated macrophages. In tumor microenvironment, a variety of stimuli (apoptotic signals, live stimulatory signals, chemo- and radio-therapy) induce tumor cells to release T-MPs, which are then taken up by monocytes, M0 or even M1 macrophages. In turn, the entered T-MPs educate monocytes, M0 or M1 macrophages into M2 like TAMs, leading to tumor growth, metastasis as well as cancer stem cell development. Data shown are representative of three independent experiments expressed as means±s.e.m. ***p < 0.001.