Figures & data
Figure 1. CD4+ T cells show cytotoxic activity against HRS cells. (A–E) Bulk (CD3), CD4+ or CD8+ T cells were isolated from PBMC of healthy donors and subsequently co-cultured with HRS cells (HL cell line L428) in a ratio of 20:1 for 14 d. Every 2–3 d T cells and L428 cells were counted by FACS analysis after staining for CD3 and CD30. The experiment was performed in triplicates and repeated with two different donors showing similar results. (A) Growth curves of HRS cells co-cultured with (w) bulk, CD4+ or CD8+ T cells in comparison to a corresponding monoculture. (B) Growth curves of bulk, CD4+ or CD8+ T cells co-cultured with HRS cells in comparison to a corresponding monoculture of unstimulated T cells. (C) Frequency of CD3-positive HRS cells (rosettes) over time. (D) Representative phase contrast images of cluster formation in 7 d-old co-cultures of HRS cells and CD4+ T cells. (E) Cluster formation was recorded by assessment of cluster diameters. The experiment was performed twice with n > 50 and in addition, HL cell lines KMH2 and L1236 were included in the cluster formation assay. (F–H) T cells and L428 cells were transplanted into NSG mice. NSG mice were first inoculated with HRS cells i.p. (F) or s.c. (G+H) and 7 d later additionally transplanted with bulk, CD4+ or CD8+ T cells. The experiment was performed with n = 5 mice per group and repeated once with similar results. (F) Mice were sacrificed after 6 weeks and analyzed for tumor burden. Representative images show i.p. tumor growth in absence of T cells. (G) Volume of s.c. tumors was measured every 3–4 d. (H) Relative (rel.) weight (% of initial weight) was assessed every 3–4 d for 6 weeks.
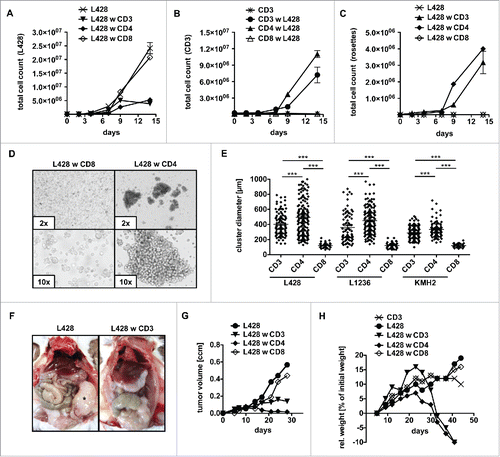
Figure 2. CD4+ T cells and HRS cells interact via immunological synapses. CD4+ T cells were isolated from PBMC of healthy donors and subsequently co-cultured with HRS cells (HL cell line L428) in a ratio of 20:1 for up to 14 d. Co-cultures were incubated either with antibodies against CD2, CD11a, CD40, CD54 and CD58 (A–B), left untreated (C) or initially treated with 75 µM dexamethasone (Dex) (D–I). (A) After 1 week, the frequency of CD3-positive HRS cells (rosettes) was assessed by flow cytometry. The experiment was performed in triplicates and repeated 3 times with similar results. (B+F) Cluster formation was recorded in parallel by phase contrast imaging to determine cluster diameter. Experiments were performed 3 times with n > 50. C, smears of co-cultures were stained for the adhesion molecule pairs CD2/CD58 (LFA-2/LFA-3) or CD11a/CD54 (LFA-1/ICAM-1) and analyzed by fluorescence microscopy. (D–E) Expression of CD2 (D) or CD11a (E) on CD4+ T cells within Dex-treated co-cultures was determined on day 7. (G-I) Every 2–3 d T cells and HRS cells were counted by flow cytometry after staining for CD3 and CD30. Experiments were performed in triplicates and repeated with two different donors showing similar results. (G) Growth curves of HRS cells co-cultured with (w) CD4+ T cells in dependence of Dex administration and in comparison to corresponding monocultures. (H) Growth curves of CD4+ T cells co-cultured with HRS cells in dependence of Dex administration and in comparison to untreated control. (I) Frequency of CD3-positive HRS cells (rosettes) over time. Experiments were performed in triplicates and repeated 3 times.
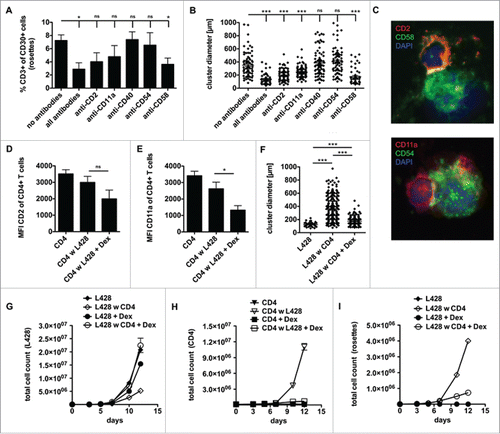
Figure 3. Anti-HRS cell activity of CD4+ T cells displays an allogeneic reaction. HLA genotype of HL cell line L428 was determined. Thereafter, CD4+ T cells were isolated from MHC-II compatible donors and subsequently co-cultured with HRS cells (A–D) or co-injected into tumor-bearing NSG mice (E, F). (A–D) HLA-matched (mCD4+) or unmatched (uCD4+) CD4+ T cells were co-cultured with HRS cells in a ratio of 20:1 for 14 d. Every 2–3 d T cells and HRS cells were counted using flow cytometry after staining for CD3 and CD30. Experiment was performed in triplicates and repeated with two different MHC-II compatible donors showing similar results. (A) Growth curves of HRS cells co-cultured with (w) mCD4+ or uCD4+ T cells in comparison to a corresponding monoculture. (B) Growth curves of mCD4+ or uCD4+ T cells co-cultured with HRS cells in comparison to untreated control. (C) Frequency of CD3-positive HRS cells (rosettes) over time. (D) Cluster formation was recorded by assessment of cluster diameters. Experiment was performed twice with n > 50. (E–G) Co-culture experiments were translated into a murine xenograft model. NSG mice were first inoculated with HRS cells s.c. and 7 d later co-injected with mCD4+ or uCD4+ T cells. Experiment was performed twice with n = 5. (E) Volume of s.c. tumors was measured every 3–4 d. (F) Relative (rel.) weight (% of initial weight) was assed every 3–4 d for 6 weeks. (G) Mice were sacrificed after 6 weeks and tumors (*) were processed by histological analysis. Representative images of s.c. tumors stained for CD3 and CD30 showing tumor-infiltration by mCD4+ T cells.
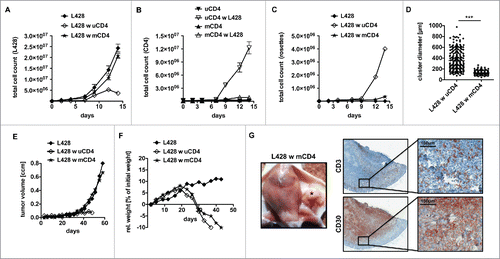
Figure 4. Gene expression profiling of co-cultures hints at interactions between MHC-II compatible CD4+ T cells and HRS cells. HLA-matched (mCD4+) or umatched (uCD4+) CD4+ T cells were co-cultured with HRS cells (HL cell line L428) in a ratio of 20:1 for 7 d. Co-cultures were separated in Dex-treated and non-treated groups. Monocultures of HRS cells served as controls. After 1 week of co-culture, HRS cells were purified by CD3-depletion and gene expression profiling (GEP) was performed, if amount of contaminating T cells was less than 5%. Controls were treated likewise. (A) Unsupervised hierarchical clustering shows differences between co-cultures and monocultures, whereas Dex-treated co-cultures clustered in one arm with monocultures. Interestingly, a random distribution of co-cultures with either mCD4+ (m) or uCD4+ (u) T cells could be observed. (B) Inflammation associated genes were significantly overexpressed by co-cultured L428 cells. (C) Gene set enrichment analysis (GSEA) showed up-regulation of genes associated with apoptosis in HRS cells after co-culture (left) and this effect was completely abolished by Dex-treatment (right). (D) Comparison of HRS cells co-cultured either with mCD4+ or uCD4+ T cells did not result in significant differences concerning apoptosis-related genes. ES, enrichment score; FDR, false discovery rate.
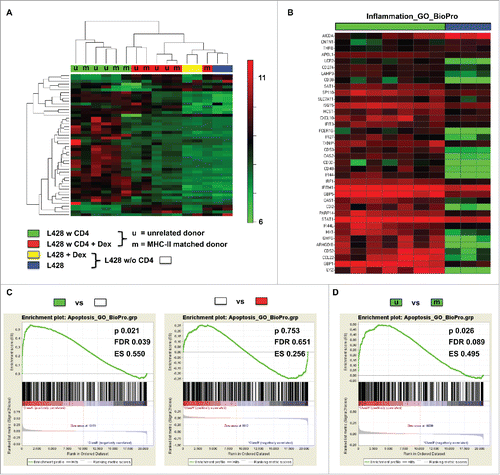
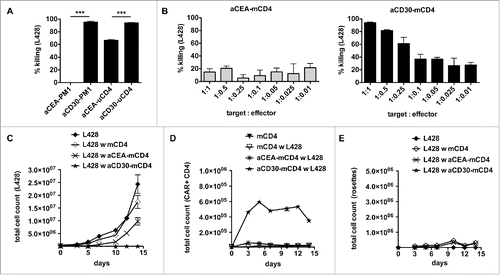
Figure 5. Gene-modified MHC-II compatible CD4+ T cells efficiently kill HRS cells. (A) TCR-negative PM1 cells were transduced with retroviral vectors encoding for anti-CEA (aCEA) or anti-CD30 (aCD30) chimeric antigen receptors (CARs) and co-cultured with GFP-expressing HRS cells (HL cell line L428). Killing efficacy was assessed after 48 h via flow cytometry. (B–E) CD4+ T cells were isolated from HLA-matched (mCD4+) or unmatched (uCD4+) donors and retrovirally transduced to express aCEA- or aCD30-CAR prior to co-culture with HRS cells. Experiments were performed in triplicates and repeated once. (B) Killing efficacy of different target (GFP-expressing HRS cells) to effector (CAR-positive CD4+ T cells) ratios was assessed after 96 h. Importantly, aCD30-mCD4+ T cells showed dose-dependent killing compared to aCEA-mCD4+ control. (C–E) Every 2–3 d T cells and HRS cells were counted by flow cytometry analysis after staining for CD3 and CAR (ratio was 1 aCD30-mCD4+ on 1 HRS cells). (C) Growth curves of HRS cells co-cultured with (w) untransduced, aCEA- or aCD30-mCD4+ T cells in comparison to a corresponding monoculture. (D) Frequency of CAR-positive mCD4+ T cells co-cultured with L428 cells in comparison to untreated or non-transduced control. (E) Frequency of CD3-positive HRS cells (rosettes) over time.