Figures & data
Figure 1. Expression of HLA-G in breast cancer correlates with outcome. Examples of immunostaining intensity: (A) no staining; (B) weak staining; (C) moderate staining and (D) strong staining used for establishing classification of the level of expression. (E, F) The Kaplan–Meier graphs was used to estimate the correlation of HLA-G expression with overall survival and relapse-free survival. Log-rank tests were used to assess significance (p values).
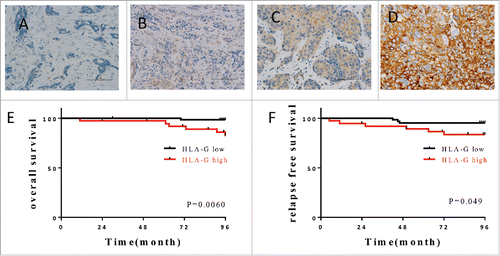
Table 1. The association of HLA-G expression with clinicopathological status was analyzed by Chi-squared tests.
Figure 2. Induction of HLA-G26–40-specific CD4+ T cell responses. (A) CD4+ T cell clones (G1; HLA-DR4/DR9/DR53, G2; HLA-DR4/DR9/DR53, G3; HLA-DR9/DR53 and G4; HLA-DR1/DR15) were tested for their capability to respond to various concentrations of HLA-G26–40 peptide using autologous PBMCs as APCs. (B) Inhibition of HLA-G26–40-specific CD4+ T cell responses by anti-HLA-DR mAb L243 but not by anti-HLA class I mAb W6/32 (negative control) (*p < 0.05, Student's t test). (C) CD4+ T cell clones were evaluated using L-cells transfected with individual HLA-DR genes as APCs to determine the restricting HLA class II alleles (*p < 0.05, Student's t test). Supernatants were collected after 48 h of incubation and analyzed by ELISA for IFNγ production (*p < 0.05, one-way ANOVA with the Holm post-hoc test). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate. (*p < 0.05, Student's t test). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate.
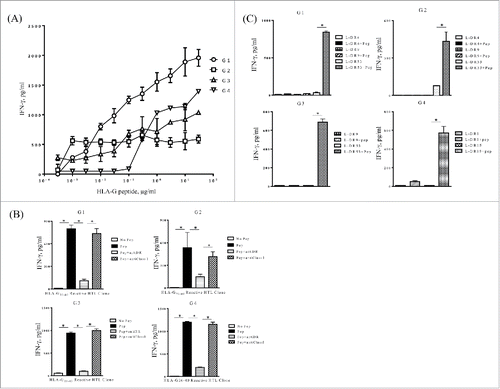
Figure 3. Evaluation of HLA-G expression in tumor cell lines after treatment with IFNγ and 5-AZA. HLA-G expression in the tumor cell lines was evaluated by Western blotting. These tumor cell lines were treated with or without IFNγ alone or in combination with 5-AZA for 72 h before tested.
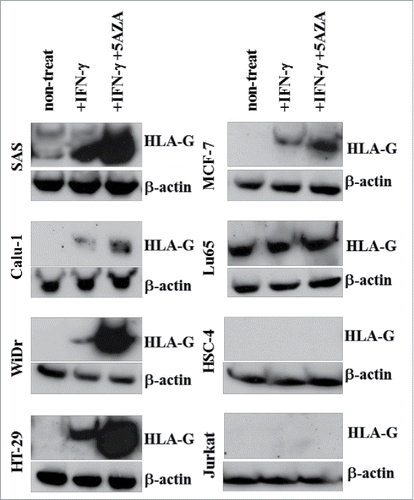
Figure 4. HLA-G26–40-specific CD4+ T cell clones directly react with tumor cells. (A) Several tumor cell lines were tested for their ability to be recognized by the HLA-G26–40-specific CD4+ T cell clones and these responses were suppressed by anti-HLA-DR mAb (*p < 0.05, one-way ANOVA with the Holm post-hoc test compared among the same tumor cell lines). (B) The HLA-G26–40-specific CD4+ T cell responses to tumor cells were enhanced by the upregulation of HLA-G molecules after treatment with 5-AZA (*p < 0.05, Student's t test). (C) The HLA-G26–40-specific CD4+ T cells reacted with naturally processed exogenous antigens presented by autologous DCs. Supernatants were collected after 48 h of incubation and analyzed by ELISA for IFNγ production (*p < 0.05, one-way ANOVA with the Holm post-hoc test compared among the same tumor cell lysate used). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate.
Figure 5. HLA-G26–40-specific CD4+ T cell clones were evaluated for their cytotoxicity against HLA-G-expressing tumor cells. The cytotoxic activity of the T cell clones (G1, G2 and G4) against HLA-DR-matched HLA-G-positive tumor cells was assessed. The HLA-G26–40-specific CD4+ T cell clones effectively lysed the tumor cells treated with IFNγ and 5-AZA. Supernatants were collected after 6 h of incubation and analyzed using a colorimetric CytoTox 96 assay. (*p < 0.05, one-way ANOVA with the Holm post-hoc test compared at the same E:T ratio among the same target).
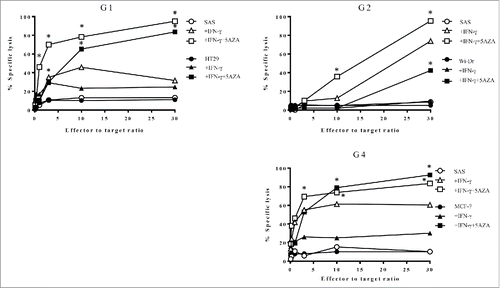
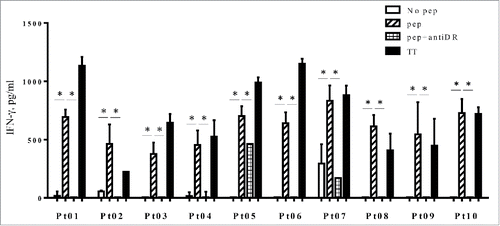
Figure 6. PBMCs from three lung cancer patients (Pt01, Pt02 and Pt03) and seven breast cancer patients (Pt04 to Pt10) were stimulated with HLA-G26–40 peptide and re-stimulated with the peptide on day 7. The supernatant of each culture was collected on day 14 and analyzed by ELISA for IFNγ production. Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate. (*p < 0.05, Student's t test). Bars and error bars indicate the mean and SD, respectively. Experiments were performed in duplicate.