Figures & data
Figure 1. IL-7 confers polyfunctionality to 6.5 TCR-Tg CD4+ T cells upon antigenic stimulation in vitro. Violet-dye-labeled spleen cells from 6.5 TCR-Tg mice were stimulated with 1 μg/mL HA peptide in the absence or presence of 100 ng/mL rhIL-7. After 7 d in culture, cells were harvested, enumerated and analyzed by FACS. (A) CD4+ T cell proliferation status as reflected by violet dye dilution. Histograms shown are representative of CD4+ T cells harvested from the specified culture conditions. Numbers indicate the percentage of divided CD4+ T cells. (B) Absolute number of CD4+ T cells recovered from the specified culture condition. Live CD4+ T cell number is calculated as: total live cells count per well × percent CD4+ T cells. Data are pooled from six independent experiments and shown as mean ± SD. (C) Expression profiles of pro-inflammatory cytokines produced by activated CD4+ T cells. Cells recovered from culture were stimulated with Leukocyte Activation Cocktail containing PMA, ionomycin and GolgiPlug for 4 h before intracellular staining for IL-2, TNFα and IFNγ. Representative dot plots shown are gated on divided CD4+ T cells, and the numbers indicate the percentage of cells in the corresponding quadrant. The results from six independent experiments are summarized in bar graph. Data are shown as mean ± SD. (D) Expression profiles of granzyme B in cultured CD4+ T cells. Histograms shown are gated on divided CD4+ T cells. Numbers in histograms indicate the percent of granzyme B-expressing cells in divided CD4+ T cells. Data are summarized in scatter plots. (E) EZH2 expression in cultured CD4+ T cells. Representative dot plots are shown to indicate EZH2 expression relative to CD4+ T-cell division. Numbers represent percent of cells in the corresponding quadrant. (F) PD1 and Foxp3 expression profiles in cultured CD4+ T cells. Representative dot plots are shown to indicate PD1 and Foxp3 expressions relative to CD4+ T-cell division. Numbers represent percent of cells in the corresponding quadrant. Summary of the results are shown in the bar graphs. Data are pooled from six independent results and shown as mean ± SD. ***, p <0 .001.
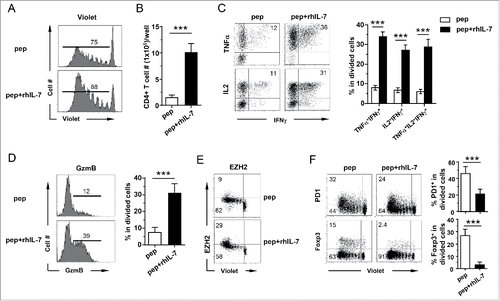
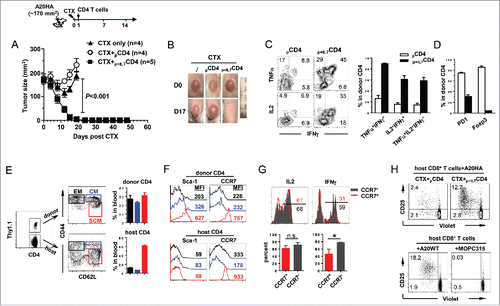
Figure 2. Polyclonal CD4+ T cells acquire polyfunctionality in the presence of rhIL-7 in vitro. Violet-dye-labeled spleen cells from a BALB/c mouse were stimulated with soluble αCD3 mAb (50 ng/mL) in the absence or presence of rhIL-7. 7 d after culture, cells were harvested and analyzed by FACS. (A) Cell proliferation status of CD4+ T cells under different culture conditions is shown in overlay histograms. (B) Expression profiles of PD1, Foxp3 and granzyme B in CD4+ T cells under different culture conditions. Results shown are representative of two independent experiments. (C) Expression profiles of pro-inflammatory cytokines produced by activated CD4+ T cells. Cells recovered from culture were stimulated with PMA and ionomycin in the presence of GolgiPlug for 4 h before intracellular staining for IL-2, TNFα and IFNγ. Representative dot plots shown are gated on divided CD4+ T cells, and the numbers indicate the percent of cells in the corresponding quadrant. (D) The results of (B) and (C) are summarized in bar graph and shown as mean ± SD. Data are pooled from two independent experiments. ***, p <0 .001.
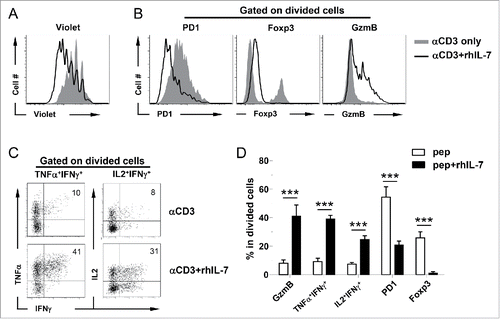
Figure 3. Induction of polyfunctionality in CD4+ T cells requires the presence of rhIL-7 during the late phase of T-cell activation. Violet-dye-labeled spleen cells from 6.5 TCR-Tg mice were stimulated with 1 μg/mL HA peptide. rhIL-7 (100 ng/mL) was added to culture following the timeline depicted in the schema. 7 d after peptide stimulation, cells were harvested and restimulated with PMA and ionomycin for 4 h before cytokine intracellular staining. Dot Plots shown are gated on divided CD4+ T cells. Data shown are representative of three independent experiments.
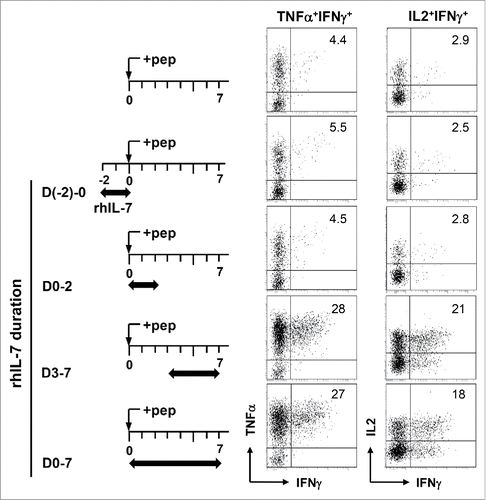
Figure 4. IL-7-induced polyfunctional CD4+ T cells exhibit increased histone acetylation in the promoters of effector cytokine genes and granzyme B but not Foxp3. (A) TSA recapitulates rhIL-7 in inducing polyfunctionality in CD4+ T cells. Spleen cells from 6.5 TCR-Tg mice were labeled with violet dye and stimulated with 1 ug/mL HA peptide. TSA (20 uM) was added to cell culture on days 2 and 3. Cells were harvested on day 4 and subjected to ICS for IFNγ, TNFα and IL-2. Dot plots shown are representative of three independent experiments. (B) Histone 3 acetylation ChIP assay. Spleen cells from 6.5 TCR-Tg mice were stimulated with 1 μg/mL HA peptide in the absence or presence of rhIL-7. 7 d later, CD4+ T cells were FACS-sorted and subjected to ChIP analysis using an antibody specific for acetylated histone 3 (H3) for immunoprecipitation. Purified naïve CD4+ T cells were included as control. The immuneprecipitated DNA was analyzed by real-time PCR to assess H3 acetylation in the promoter regions of IFNγ, TNFα, IL-2, granzyme B and Foxp3. Results are normalized to input DNA. Data shown are representative of two independent experiments with similar results. *, p <0 .05; **, p <0 .01.
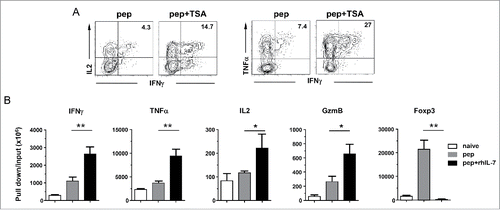
Figure 5. IL-7-driven polyfunctionality is STAT5-dependent. The experimental timeline is depicted in the schema. Spleen cells from DO11.10 TCR-Tg mice were labeled with violet dye and stimulated with the cognate peptide for 24 h. Cells were then harvested and spun infected with CA-STAT5 or DN-STAT5 retrovirus carrying the Thy1.1 marker. 12 h after infection, cells were harvested and restimulated with peptide-pulsed spleen cells from CD45.1 mice as APCs with or without addition of rhIL-7 (100 ng/mL). Cells were subsequently cultured for an additional 4 d and subjected for FACS analysis. (A) Viral transduction efficiency in CD4+ T cells evaluated by expression of Thy1.1 marker. Histograms shown are representative of three independent experiments. Numbers indicate the percent of virus-infected cells in total CD4+ T cell population. (B) Cytokine profiles of virus-infected CD4+ T cells. Cells recovered from culture were subjected to intracellular staining for TNFα and IFNγ. Representative dot plots shown are gated on divided CD4+ T cells. Numbers indicate the percent of TNFα+ IFNγ+ cells. (C) Summary of the results shown in (B). (D) Expression profiles of Foxp3 in virus-infected CD4+ T cells. Numbers indicate the percent of Foxp3-positive cells in divided CD4+ T cells. (E) Summary of the results shown in (D). Data are pooled from three independent experiments and shown as mean ± SD. (F) Histone 3 acetylation ChIP assay. Spleen cells from 6.5 TCR-Tg mice were transduced with CA-STAT5 retrovirus in the absence of rhIL-7. Infected CD4+ T cells were FACS-sorted and subjected to H3ac ChIP assays. CD4+ T cells stimulated with peptide in the absence or presence of rhIL-7 were used for comparison. The immuneprecipitated DNA was analyzed by real-time PCR to assess H3 acetylation in the promoter regions of IFNγ, TNFα and Foxp3. Results are normalized to input DNA. Data shown are representative of two independent experiments with similar results. *, p <0 .05; **, p <0 .01; ***, p <0 .001. n.s., not statistically significant.
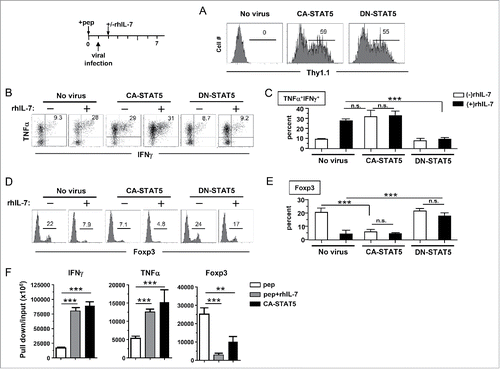
Figure 6. In vitro-generated polyfunctional CD4+ T cells expand intratumorally in response to immunization. (A) Adoptive transfer of rhIL-7-conditioned polyfunctional CD4+ T cells followed by vaccination exerts robust antitumor effects in vivo. The schema outlines the timeline of experimental procedures. Mice bearing three-day-old subcutaneous A20HA-luci tumors were not treated or treated as indicated. Tumor growth curves of each group are shown. Results are presented as mean ± SD of tumor area. Mice were also subjected to BLI periodically to visualize the tumor burden. Representative images of mice in each group before and after treatment are shown in (B). (C) Intratumoral expansion of the transferred polyfunctional CD4+ T cells in response to immunization. Following the procedures depicted in the schema, luciferase-transduced polyfunctional CD4+ T cells (p+IL7CD4+-luci) were transferred to tumor-bearing mice followed by PBS or vacHA injection. BLI was conducted to detect luciferase-tagged donor CD4+ T cells. Representative images of mice in each group at specified time are shown. Results of T-cell luciferase signal intensity quantified as photon/sec are summarized in (D). Each symbol in plot represents one mouse. The lines represent the average value of T-cell signal intensity.
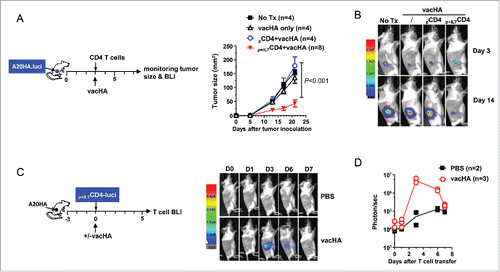
Figure 7. Adoptive transfer of polyfunctional CD4+ T cells following lymphodepletive chemotherapy leads to complete regression of large tumors, accompanied by antigen epitope spreading and persistence of stem cell-like memory T cells. The schema outlines the timeline of experimental procedures. Mice were inoculated with A20HA subcutaneously in the flank. When tumor sizes reached ∼170 mm2, mice were treated with CTX (150 mg/kg). The next day, some mice received adoptive transfer of in-vitro-activated, non-polyfunctional HA-specific CD4+ T cells (pCD4+), while some other mice received IL-7-conditioned polyfunctional HA-specific CD4+ T cells (p+IL7CD4+). (A) Tumor growth curves of mice in each group are shown. Results are presented as mean ± SD of tumor area. (B) Representative images of tumors in mice under each condition before and after treatment. (C) Polyfunctional status of the transferred CD4+ T cells. Tail blood was collected from mice 17 d after T-cell transfer. Leukocytes in blood samples were stimulated with PMA/ionomycin in the presence of GolgiPlug for 4 h before cytokine ICS. The cytokine profiles of the donor CD4+ T cells, revealed by FACS, are shown by representative dot plots. Bar graph summarizes the percent of donor CD4+ T cells capable of simultaneously producing two or three cytokines. (D) Percent donor CD4+ T cells expressing PD-1 or Foxp3. Results are presented as mean ± SD of at least three samples per group. (E) Persisting donor CD4+ T cells exhibit features of Tem, Tcm and Tscm cells. Tail blood from cured mice receiving CTX+p+IL7CD4+ was collected and examined for expressions of CD44, CD62L, Sca-1 and CCR7 in donor and host CD4+ T cells. Three subsets are gated based on distinct CD44CD62L patterns. Percent of the gated populations is summarized in bar graph. Expression patterns of Sca-1 and CCR7 of the gated cells are shown in (F). Numbers in histogram represent the values of MFI of Sca-1 or CCR7. (G) Cytokine expression relative to CCR7 in donor CD4+ T cells. Leukocytes from the peripheral blood of cured mice were stimulated with PMA/ionomycin and subjected to ICS to reveal the relation of IL-2 and IFNγ to CCR7 in donor CD4+ T cells. Numbers represent percent of cytokine-producing cells in CCR7+ or CCR7− donor CD4+ T cells. (H) CTX+p+IL7CD4+ treatment leads to tumor antigen epitope spreading and activation of host CD8+ T cells. 20 days after T-cell transfer, mice treated with CTX+pCD4+ or CTX+p+IL7CD4+ were sacrificed. Purified splenic CD8+ T cells were labeled with violet dye and incubated with irradiated A20HA, A20WT or MOPC315 cells. 7 d later, CD8+ T cells were harvested and examined for proliferation/activation by evaluating violet-dye-dilution and CD25 expression. ***, p <0 .001.