Figures & data
Figure 1. Macrophages contribute to mesothelioma tumor growth. C57BL/6J mice were inoculated s.c. with AE17 mesothelioma tumor cells (3 × 105 cells) or co-inoculated with BM-derived macrophages (1 × 105 cells) and AE17 cells (3 × 105 cells, ratio 1 macrophage:3 tumor cells) and tumor growth monitored (). In a separate experiment, mice were inoculated with 5 × 105 AE17 tumor cells s.c and tumors left to grow to 25–30 mm2 before anti-F4/80 Ab treatment commenced (). Daily anti-F4/80 Ab injections (100 μg/mL in 100 µL) were alternately given i.p. then i.t. for 9 d. Control mice were given the PBS diluent. Pooled data is shown as mean ± SEM; n = 9 mice/group. Peritoneal macrophages from C57BL/6J mice were cultured overnight with IFNγ/LPS (M1 stimuli), IL-4/IL-13 (M2 stimuli), 50% AE17 mesothelioma-derived supernatant or left untouched (no stimuli, NS) and analyzed by flow cytometry for surface expression of CX3CR1 (), CD206 (), and CD40 (G and H). Pooled data from four experiments is shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
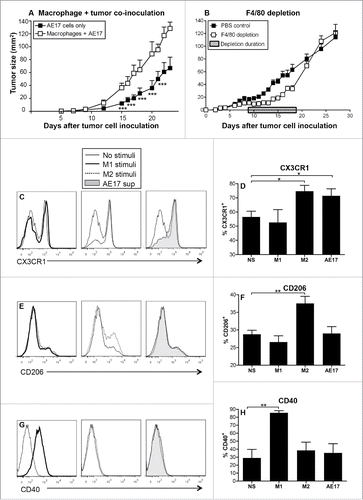
Figure 2. Mesothelioma tumor-derived factors polarize macrophages into M2-like cells. Peritoneal macrophages from C57BL/6J mice were cultured overnight with IFNγ/LPS (M1 stimuli), IL-4 (M2 stimuli), 50% AE17 mesothelioma-derived supernatant or left untouched (no stimuli, NS). Supernatants were analyzed for TGF-β (2A) and MCP-1 () by CBA. AE17 mesothelioma supernatant contains TGF-β (438 ± 194 pg/mL) and MCP-1 (4658 ± 136 pg/mL), therefore the data shown for AE17-exposed macrophages in were calculated by subtracting AE17 supernatant only. CFSE-labeled Balb/c non-adherent splenocytes were added to macrophages at varying ratios. After 5 d, cells were stained for CD8+ T cells and analyzed by flow cytometry. The percentage of CD8+ T cell proliferation was calculated based on loss of CFSE staining intensity of the parent peak (example histograms and graphed data from four experiments in ). Supernatants from the macrophage:T cell ratio of 1:2 MLR assay for IFNγ by CBA (), the data shown was calculated by subtracting the IFNγ levels seen in the M1 controls (i.e., 54.3 ± 16.1 pg/mL) where appropriate. Pooled data from four experiments is shown as mean ± SEM. *p < 0.05, **p < 0.01.
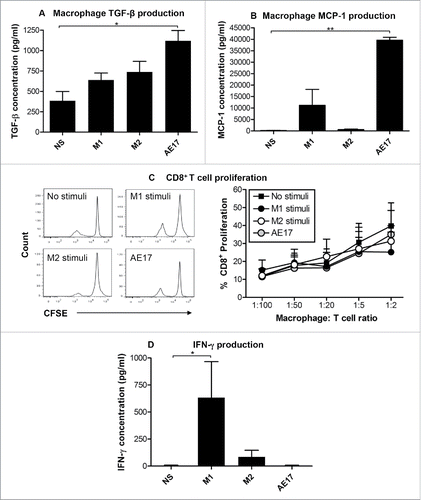
Figure 3. MDSCs dominate the spleens and bone marrow of mesothelioma-bearing mice. C57BL/6J mice were inoculated s.c. with 5 × 105 AE17 tumor cells. Spleens and BM collected from mice with small (15.4 ± 2.8 mm2) or large (113.4 ± 8.4 mm2) tumors were stained for macrophage subsets and MDSCs. After gating on CD11b+F4/80+ macrophages () CD11b+F4/80+Ly6ChiCX3CR1lo M1 cells and CD11b+F4/80+Ly6CloCX3CR1hi M2 cell were identified. Gating on CD11b+F4/80lo cells was used to identify spleen and BM MDSCs (CD11b+F4/80loLy6C+Ly6G+; ). Data from healthy control mice (n = 10); small tumor-bearing (n = 20); and large tumor-bearing mice (n = 12) shows CD11b+F4/80+ macrophages in spleens () and BM (), splenic CD11b+F4/80+ macrophage intracellular IL-10 production (, n = 5–7/group), as well as splenic and BM macrophage subsets (, respectively). Data is shown with each point representing an individual mouse, the line shows the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
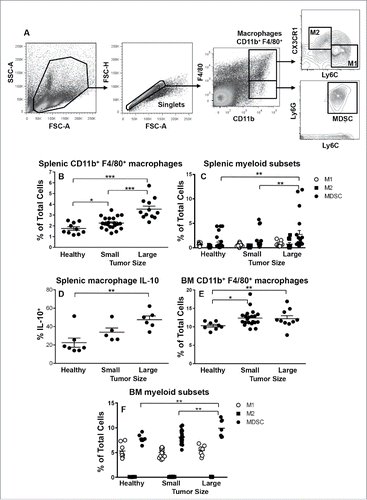
Figure 4. M3 macrophages dominate the mesothelioma tumor microenvironment. Small or large tumors were stained for macrophage subsets and MDSCs to identify M1 cells, M2 cells and M3 macrophages (CD11b+F4/80+Ly6ChiCX3CR1hi) and MDSCs. The gating strategy () demonstrates gating on CD11bhiF4/80hi macrophages as there were no F4/80low macrophages or F4/80loCD11b+Ly6G+MDSCs. Data from small tumor-bearing mice (n = 25) and large tumor-bearing mice (n = 21) for CD11b+F4/80+ macrophages (), macrophage subsets () and tumor-associated macrophage subset IL-10 (, n = 8/group) and TNF-α (, n = 8/group). Data is shown with each point representing an individual mouse, line shows the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
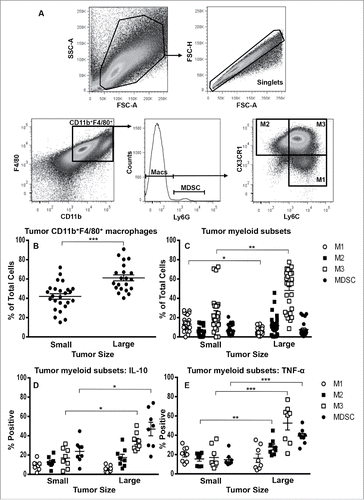
Figure 5. M3 cells expand in tumors during disease progression. Tumor-associated cells were first stained for M1, M2, M3 and MDSCs as described for and then stained with Ki67 or DAPI for cell cycle analysis (, respectively). Data from AE17-tumor-bearing mice (n = 5–6/group) show the proportions of M1, M2, M3 cells and MDSCs that were in G2 mitosis phase (). Data is shown with each point representing an individual mouse, the line shows the mean ± SEM; ***p < 0.001, ****p < 0.0001. Immunofluorescence on large frozen AE17 mesothelioma tumor sections (n = 3) stained with F4/80 (red), CX3CR1 (green) and Ly6C (blue) revealed micro-niches of M1 and M3 cells (). Exposure times were determined by comparing each fluorescence stain to the relevant isotype control; a representative photograph is shown, magnification 200×.
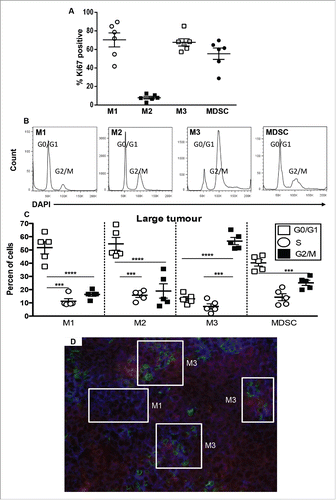
Figure 6. Gemcitabine does not affect suppressive M2 cells in spleens and tumors. C57BL/6J mice were inoculated with 5 × 105 AE17 tumor cells s.c. and tumors left to grow to 15–20 mm2 before gemcitabine treatment commenced. Treatment consisted of three i.p. injections (120 µg/g of body weight) every 3 d (). Pooled data is shown as mean ± SEM, n = 11 mice/group. In a separate experiment, AE17-bearing mice were given three injections of PBS (100 µL/dose) or gemcitabine every 3 d. Tumors (), spleens () and DLNs () were collected 4 d after the last dose, stained for CD11b+F4/80+ macrophages, M1, M2 or M3 subsets and MDSCs and analyzed by flow cytometry. Data is shown with each point representing an individual mouse (n = 5 mice/group), the line shows the mean ± SEM. *p < 0.05, **p < 0.01.
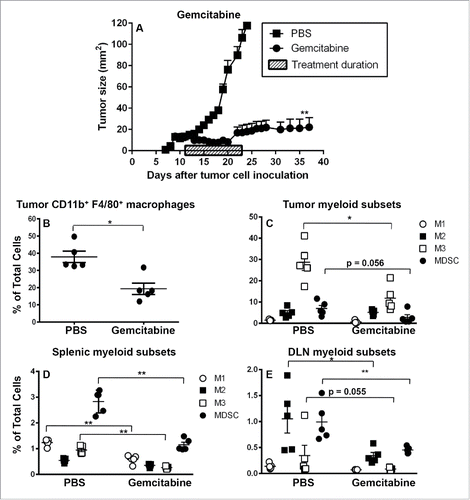
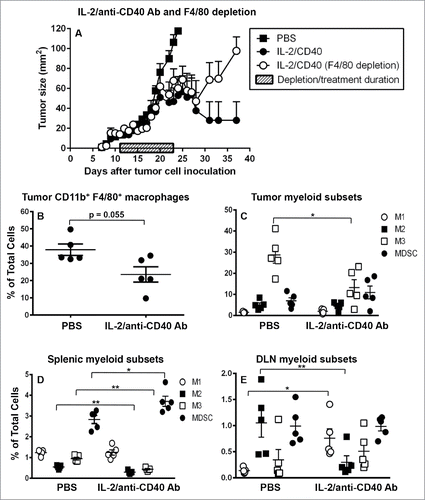
Figure 7. IL-2/anti-CD40 Ab promotes M1 cells in draining lymph nodes. C57BL/6J mice were inoculated with 5 × 105 AE17 tumor cells s.c. and tumors left to grow to 15–20 mm2 before anti-F4/80 Ab injections commenced. IL-2/anti-CD40 Ab treatment consisted of three i.t. injections (20 µg IL-2 and 40 µg anti-CD40 Ab, 100 µL/dose) given every 2 d (). Daily anti-F4/80 Ab injections were given i.p. and i.t. alternately for 9 d. Pooled data shown as mean ± SEM, n = 7–10 mice/group. In a separate experiment, AE17 tumor-bearing mice were given three doses of either PBS (100 µL/dose) or IL-2/anti-CD40 Ab i.t. every 2 d and samples collected 3 d after the final dose. Tumors (), spleens (, and DLNs () were stained for CD11b+F4/80+ macrophages, M1, M2 or M3 cells and MDSCs for analysis by flow cytometry. Data is shown with each point representing an individual mouse (n = 5 mice/group), the line shows the mean ± SEM. *p < 0.05, **p < 0.01.