Figures & data
Figure 1. Suppressive Tregs accumulate at T site in CRC patients. (A–B) Representative CD127/FOXP3 staining (A) and percentages of Tregs (B) in gated CD4+ T cells in PB, NT and T samples of CRC patients (n = 31). *p < 0 .05, ****p < 0 .001, by Wilcoxon matched-pairs test, two-tailed. In all figures, Tregs and Tconvs have been identified as FOXP3+ CD127low or FOXP3−, respectively, within the CD14− CD16− CD56− CD19− viability dye− CD4+ gate. (C–E) CFSE profile overlay (C), mean fluorescence intensity (MFI) of CFSE (D), and percentage of inhibition of CFSE dilution (E) in gated Tconvs (CD4+FOXP3−) or CD8+ T cells from Treg-depleted or total (Ctrl) mononuclear cells, obtained from a representative CRC patient. The experiment was repeated thrice with cells of individual patients giving similar results. *p < 0 .05, **p < 0 .01, ***p < 0 .005, by Student t test, unpaired.
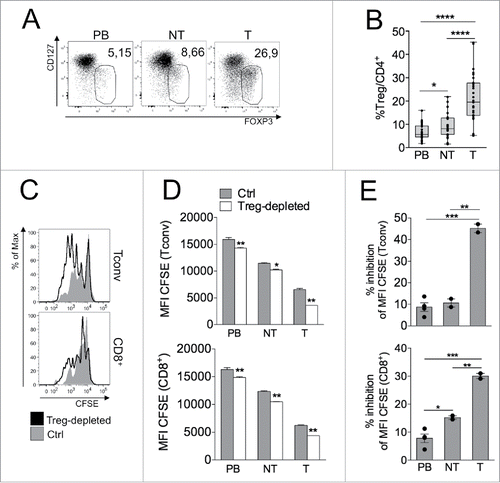
Figure 2. Helioshigh, epigenetically stable, Tregs specifically increase in tumor. (A–B) Representative Helios/FOXP3 staining in overlaid Helioshigh Tregs (red), Helioslow Tregs (blue) and Tconvs (gray) (A) and percentages of Helioslow and Helioshigh Tregs in gated CD4+ T cells (B) in PB, NT and T of CRC patients (n = 21). **p < 0 .01, ***p < 0 .005, ****p < 0 .001, by Wilcoxon matched-pairs test, two-tailed. ns, not significant. (C) The indicated subsets were FACS-sorted from PB and NT samples of a CRC patient and TSDR analysis was conducted as detailed in the Patients and Methods section. We could not perform TSDR analysis from any T specimen due to a very low yield of recovered cells. Color-coding represents the methylation status; numbers on the top indicate the sequential position of each CpG residue; percentages on the right refer to the frequency of demethylated sites.
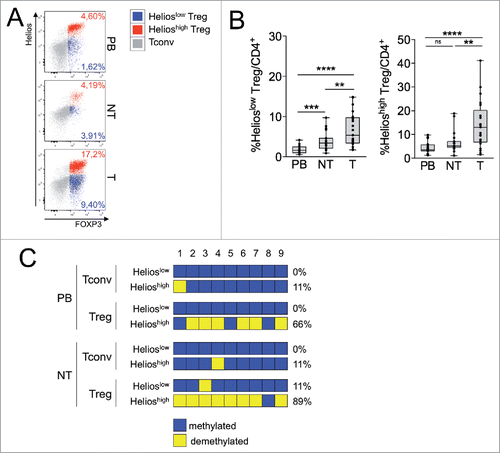
Figure 3. T-Tregs express OX40, conveying proliferative signal, at highest levels. (A–C) Representative CD45RA/OX40 staining (A) and OX40+ percentages in gated Tregs and Tconvs (n = 29) (B) or Helioshigh vs. Helioslow Tregs (n = 20) (C) from PB, NT and T of CRC patients. *p < 0 .05, **p < 0 .01, ***p < 0 .005, ****p < 0 .001, by Wilcoxon matched-pairs test, two-tailed. (D–E) Spearman correlations between the percentages of OX40+ Tconvs and Tregs (D) and of OX40+ Helioshigh or Helioslow Tregs (E). *p < 0 .05, **p < 0 .01, ****p < 0 .001. (F) Mononuclear cells, obtained from the T sample of a representative CRC patient, were stimulated in the presence of scaled concentrations of rOX40L, and proliferation was estimated in gated CD8+, Tconvs (CD4+FOXP3−) and Tregs (CD4+FOXP3+), in terms of the relative change in CFSE MFI. The experiment was independently repeated in two patients giving similar results. *p < 0 .05, **p < 0 .01, by Student t test, unpaired, compared to ctrl.
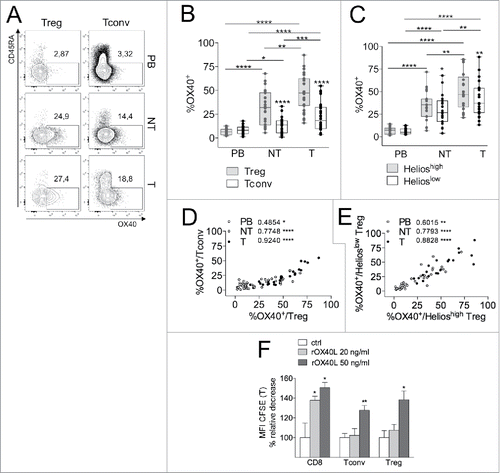
Figure 4. Both genetic and microenvironmental factors contribute to set high CD39 levels in T-Tregs. (A) Representative histogram overlay of CD39 profile in total lymphocytes (gray areas), gated Tconvs (black lines) or gated Tregs (dotted lines) in the PB of CRC patients, on the basis of their genotype at the SNP rs10748643 (AA, AG or GG). (B) Percentages of CD39high cells in gated Tregs and Tconvs from PB, NT and T samples of CRC patients, stratified into AA (n = 9), AG (n = 9) or GG (n = 3) genotypes. *p < 0 .05, **p < 0 .01, ***p < 0 .005, ****p < 0 .001, by Student t test, paired, between Tregs and Tconvs, by Wilcoxon matched-pairs test between different districts, and by Student t test, unpaired, between different genotypes. (C) Concatenated analysis of Helios, CD39 and OX40 expression in different subpopulations (Tregs and Tconvs), compartments (PB, NT and T) and genotypes (AA, AG and GG). Each pie represents grouped data from seven (AA), eight (AG) or two (GG) patients.
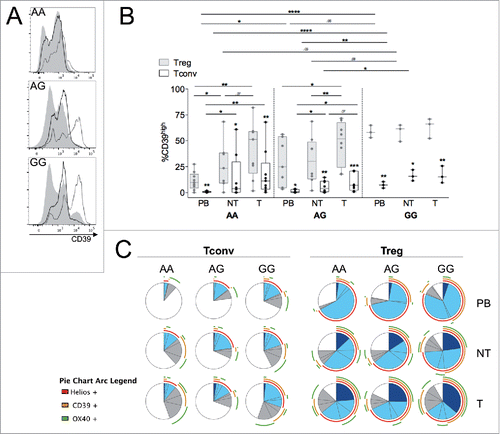
Figure 5. Th17-like Tregs, contained in the CD39highHelioslow subset, accumulate at T site. (A) Percentages of IL-17-producing Tregs and Tconvs in different compartments from CRC patients (n = 14). *p < 0 .05, **p < 0 .01, ***p < 0 .005, by Student t test, paired, between Tregs and Tconvs, or by Wilcoxon matched-pairs test between different districts. (B–C) Spearman correlations between the percentages of IL-17+ Tconvs and Tregs (B) and of IL-17+ Tconvs respect to total Tregs (C). *p < 0 .05; ns, not significant. (D) Cytokine amounts (estimated as the pg released in 18 h per gram of tissue) in NT- compared to T-CM from CRC patients (n = 19). **p < 0 .01, by Wilcoxon matched-pairs test. (E) Representative IL-17 versus CD39 expression in Tregs or Tconvs from the indicated districts. (F) Representative IL-17 vs. Helios (left) or OX40 (right) expression in T-Treg and T-Tconv subsets distinguished on the basis of CD39 and IL-17 expression. Data shown are representative from one out of four patients tested.
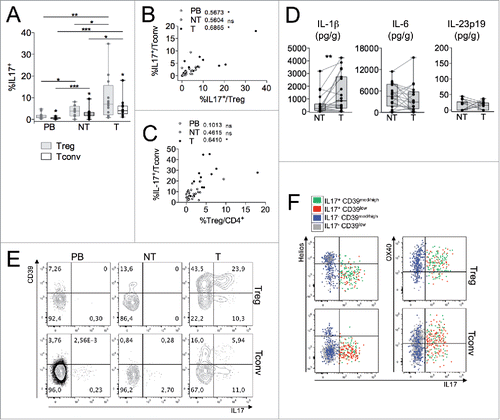
Figure 6. Tfh and Tfr, enriched in the CD39lowOX40+ subset, expand in both NT and T compartments. (A–B) Representative CXCR5/BCL6 staining (A) and percentage of BCL6+CXCR5+ follicular cells (B) in gated Tregs and Tconvs from PB, NT and T of CRC patients (n = 16). *p < 0 .05, ***p < 0 .005, by Student t test, paired, between Tregs and Tconvs, or by Wilcoxon matched-pairs test between different districts. (C) Overlay of the indicated markers in the total Treg or Tconv pool (gray areas) compared to the T follicular subpopulation (black lines), in a representative T sample from three patients analyzed.
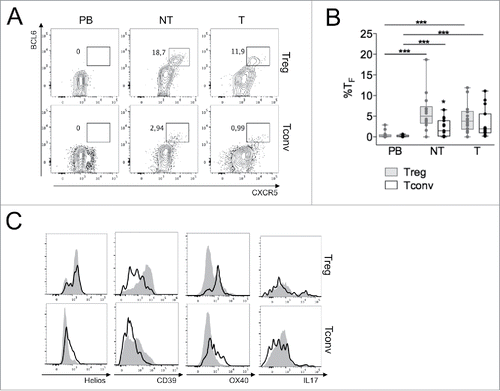
Table 1. Characteristics of all CRC patients included in the study (n = 34 , not treated with any neoadjuvant therapy).
