Figures & data
Figure 1. Computational approaches to identify cancer-specific MHC ligands. Comparing exome sequencing data (A) from tumor and healthy tissues allows identification of non-synonymous mutations. From mRNA sequencing (B) cancer-specific gene fusions, aberrant RNA specific, and expressed/overexpressed TA genes can be identified by comparison with databases of splice isoforms and healthy tissues. All possible peptides (blue lines) containing at least one cancer-specific amino acid mutation are then subjected to MHC-binding peptide predictions. For MHC I molecules 8- to 11-mer and for MHC II 14- to 20-mer peptides are considered for predictions. Predicted peptides are ranked based on their predicted binding affinity and possibly other variables, like immunogenicity, pMHC complex stability, predicted cleavage sites or level of expression of the corresponding genes.
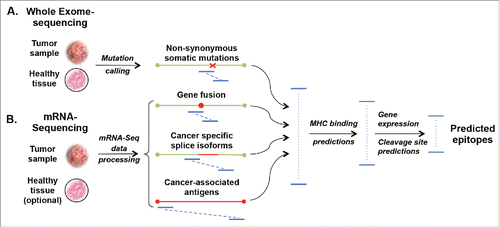
Figure 2. MHC peptide binding assays. (A) Widely used MHC I peptide binding assays include the following: (i) Rebinding of test peptide (red dot) and β2m (brown dot) after acid strip of immobilized pMHC containing an irrelevant peptide (yellow dot). The newly formed complexes are quantified by means of a conformational mAb (green Y) containing a fluorescent label (blue star). (ii) Rescuing with test peptides (blue dot) of UV irradiated complexes containing a photocleavable peptide. The newly formed complexes can be captured and quantified or used for preparing combinatorial multimer libraries. (iii) Test peptide-driven refolding of β2m and biotinylated MHC I heavy chain is quantified by a luminescent oxygen channeling immunoassay reporting the proximity of an pMHC bearing acceptor bead and an acceptor bead coated with conformational pan-MHC I mAb. (B) Peptide-MHC class II binding assay using peptide microarray. Peptides (15 mer) (colored dots) synthesized on a microarray chip and incubated with receptive soluble MHC II molecules (blue dimers) and after washing peptide bound MHC II are quantified by means of a fluorescently labeled (blue star) conformational mAb (green Y) as described for A.
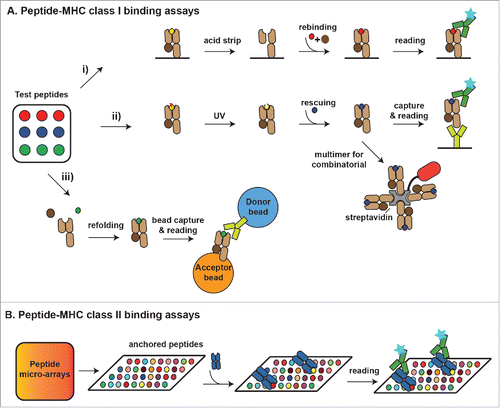
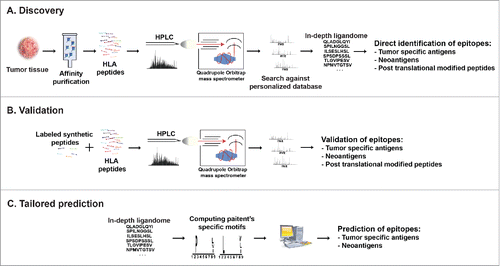
Figure 3. Cancer peptidome analysis. (A) Overview of immunopeptidome MHC ligand discovery. HLA molecules are isolated from cells, their peptide cargo isolated and analyzed by LC-MS. Matching the MS data to customized reference databases that include information of somatic mutations allows direct identification of MHC ligands derived from TA including those containing post-translational modifications. (B) For validation and quantification synthetic peptides labeled with heavy isotopes spiked into the eluted peptidome sample permit validation of in vivo presented MHC ligands. (C) Patient's-specific HLA peptide binding motifs can be resolved from in-depth ligandome data sets and may be used to predict private tumor-specific MHC ligands.