Figures & data
Figure 1. Breast cancer cell lines do not express TSLP mRNA. (A) TSLP mRNA expression quantified by quantitative PCR (TaqMan) in 11 breast cancer cell lines. PBMC and MRC5 correspond to negative and positive control respectively. N = 4. (B, C, D) Boxplots in the left panels represent mRNA expression in 1036 cancer cell lines from CCLE of TSLP, IL-8 and SDF1 respectively. (B, C, D) Boxplots in the right panels show the gene expression of TSLP, IL-8 and SDF1 respectively for breast cancer cell lines, which were grouped according to their corresponding molecular subtype. p values were calculated with a t test comparing different cancer cell subtypes.
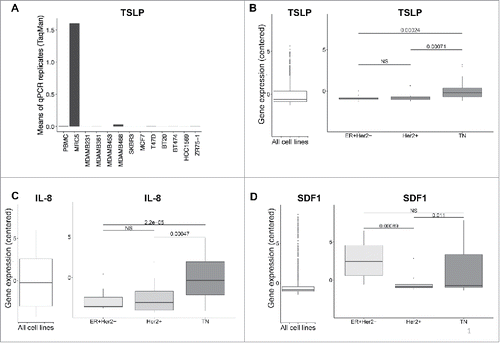
Figure 2. TSLP mRNA level is higher in normal breast than breast cancer tissue. (A, B, C) mRNA expression in 58 breast cancer patients from TCGA of TSLP, IL-8 and SDF1 respectively. Boxplots represent data of tumors classified in three different subtypes, namely Luminal (ER+Her2−), Her2+ and Triple negative (TN) and normal breast tissue as indicated. p values were calculated with a t test comparing different clinical groups.
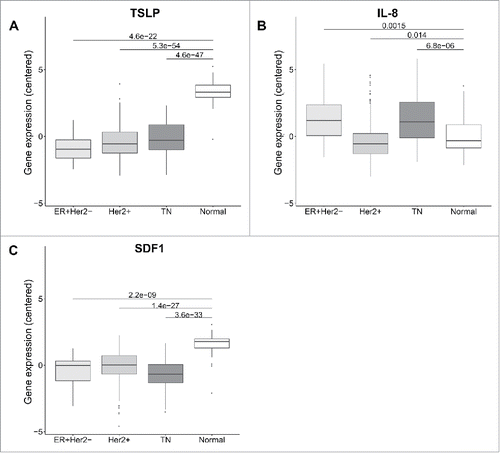
Figure 3. TSLP mRNA expression is higher in juxta-tumor tissue than breast cancer tissue. (A) TSLP transcripts were measured by quantitative PCR (TaqMan) in 19 breast cancer tissues (black bars) and 19 corresponding juxta-tumor tissues (gray bars). White bars represent TSLP levels detected in MRC5 and PBMC used as positive and negative control respectively. (B) Quantification of TSLP mRNA transcripts shown as percentage of housekeeping gene expression. Four housekeeping genes were used for these experiments: Actin Beta (ACTB), Hypoxanthine Phosphoribosyltransferase 1 (HPRT1), Ribosomal Protein L31 (RPL31) and Beta-2-Microglobulin (B2M). Lines represent mean +/− the Standard Error of the Mean (SEM). Wilcoxon matched pairs test was used to calculate p value. N = 19. (C) Quantification of soluble TSLP measured by ELISA in the supernatants generated from primary breast tumor tissues and corresponding juxta-tumor samples as described in the Material and Methods section. ELISA sensitivity detection limit, which is represented by the dashed line, was 31 pg/mL as recommended by the manufacturer instructions. N = 40.
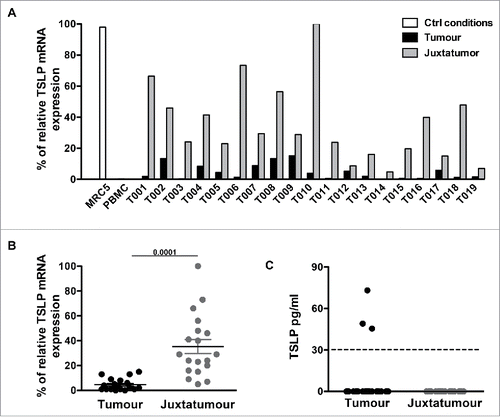
Figure 4. Breast cancer tissues do not express TSLP. (A) Tonsil sections were used as positive control tissue to validate TSLP staining by immunohistochemistry. TSLP and matched isotype staining was performed in two consecutive tonsil sections. Arrows indicate positive staining. (B) Isotype, TSLP and H&S staining in three consecutive slides of four representative breast cancer specimens. Arrows indicate positive staining. All tissue sections are shown at 20x and 40x magnification. (C) Percentages of TSLP staining in epithelial cells. Each symbol represents a different sample.
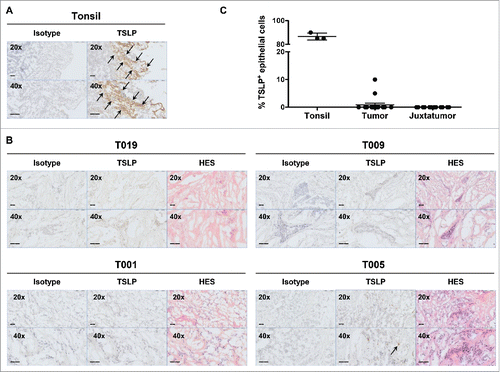
Figure 5. TSLP receptor is not detected in primary breast cancer by flow cytometry. (A–B) Flow cytometry dot plots from one representative human primary breast tumor. (A) Gating strategy used to investigate the expression of TSLP-R and IL-7-Rα in viable CD45+Lin− (Q1), CD45+Lin+ (Q2) and CD45−Lin− (Q4). (B) Gating strategy designed to detect DC in primary breast cancer. DC were defined as DAPI−CD45+Lin−HLA-DRhighCD11chigh. The expression of TSLP-R and IL-7-Rα was assessed in DC (Q6) as well as HLA-DRhighCD11c− (Q5) and HLA-DR−CD11c− (Q8). (C) Percentages of TSLP-R+IL-7-Rα+ DC in tumors, corresponding juxta-tumor tissues and in the dermis of normal skin. Lines represent mean +/− the Standard Error of the Mean (SEM). N = 6 (D) TSLP-R and IL-7-R expression in DC enriched from PBMC without digestion step (gray histogram, Undig) and after mechanical and enzymatic digestion (dark gray histogram, Dig), left panels. One representative background histogram staining is shown in light gray. Quantification of TSLP-R+IL-7-Rα+DC in undigested and digested enriched DC is shown in the left panel. N = 3.
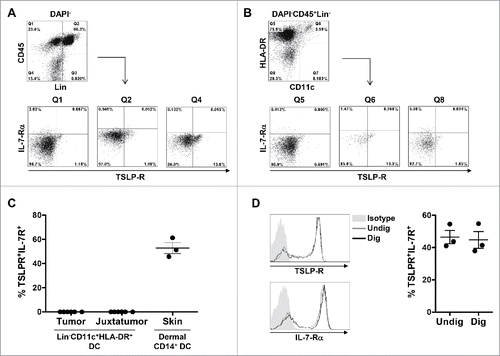
Table 1. Clinical information of patients included in the study.
