Figures & data
Figure 1. Detection of DR11/AE37 CD4+ T cells in peptide-stimulated cultures from PBMCs of DRB1*11+ prostate cancer patients following vaccination. (A) PBMCs from patient no.10 (PR10), isolated from a blood sample collected during vaccinations (after 1st vaccine inoculation; R2), were assessed with DR11/AE37 tetramer following in vitro AE37-stimulation and analyzed by flow cytometry. According to the representative example of the gating strategy depicted, DR11/AE37 tetramer+ cells were identified in CD4+ T lymphocytes, after exclusion of doublets, dead, and generally non-specifically binding cells. An FMO (fluorescence minus one) control was used to determine tetramer positivity. Final dot plot presents DR11/A37+ T cells gated on total alive CD4+ T cells (after exclusion of CD14+CD16+CD19+cells). (B) Pre-vaccination, during and post-vaccination PBMCs' samples from HLA-DRB1*11+ patients, were stimulated in vitro with AE37 peptide, followed by DR11/AE37 tetramer and surface mAbs staining and analyzed by flow cytometry. Graph presents proportions of DR11/AE37+CD4+ T cells detected in all patients analyzed at pre-vaccination, during and post-vaccination, at the timepoints indicated. (C) Dot plots and proportions of DR11/AE37+ CD4+ T cells, from patients PR14 and PR15, detected in AE37-stimulated cultures from blood samples collected at the indicated timepoints.
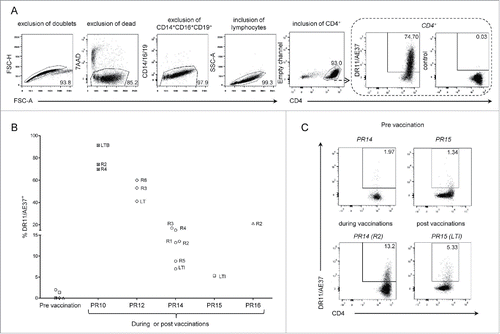
Figure 2. Detection of vaccine-specific memory CD4+ T cells. (A) Assessment of total number of Central memory-CM (CD45RA−CCR7+CD28+) and Effector memory-EM (CD45RA−CCR7−CD28+) CD4+ T cells before (ex vivo) and after AE37 peptide in vitro stimulation, from DRB1*11+ vaccinated patients. (B) Gating strategy for identification of CM and EM CD4+ T cells in AE37 in vitro stimulated samples after vaccination. Alive CD4+ T cells, after exclusion of CD14+CD16+CD19+ cells were gated according to CD45RA and CCR7 expression. Subsequent gating based on CD28 expression identified CM and EM cell subsets, which were analyzed for vaccine specificity using DR11/AE37 tetramer. Dot plots of PR14 with samples collected 4 y post-vaccinations are presented. (C) Shown are percentages of CM and EM DR11/AE37+ CD4+ T cells for all patients analyzed.
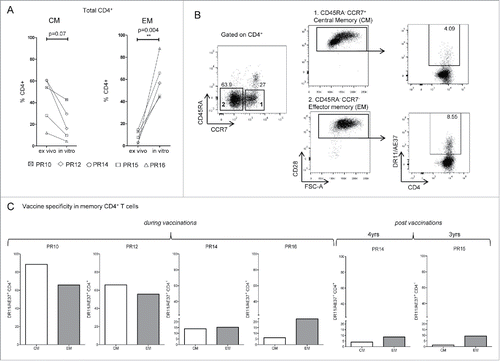
Figure 3. Assessment of DR11/AE37 CD4+ T cells in naive and memory subsets. (A) Purified naive and memory CD4+ T cell populations were isolated from circulating peripheral lymphocytes, from patient PR14, 4 y post-vaccinations by FACS cell sorting. Sorting strategy included separation of three defined CD4+ T cells subsets: (1) naive (CD45RA+CCR7+CD28+) and memory subsets (2) CM (CD45RA−CCR7+CD28+) and (3) EM (CD45RA−CCR7−CD28+). (B) Sorted populations were stimulated in vitro with the AE37 peptide and assessed at day 8 with DR11/AE37 tetramer as in .
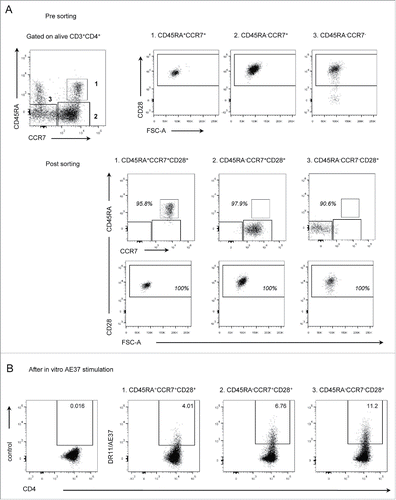
Figure 4. Functional profile of AE37-responsive CD4+ T cells. (A) After 10 d of in vitro stimulation, AE37-expanded CD4+ T cells were restimulated for 16 h with AE37 peptide-loaded DCs and cytokine/GzB production was assessed with intracellular staining. Unloaded DCs were used for determination of background cytokine and GzB production. An example of dot plots from PR16 presents the percentages of cytokine or GzB producing CD4+ T cells. (B) Data summary from for all patients analyzed. Blood samples were collected during (PR10, PR12, PR16) and post (PR14, PR15) vaccinations. (C) Cumulative results (%) (Median values) of CD4+ T cells producing the indicated cytokines, from all patients analyzed, are shown.
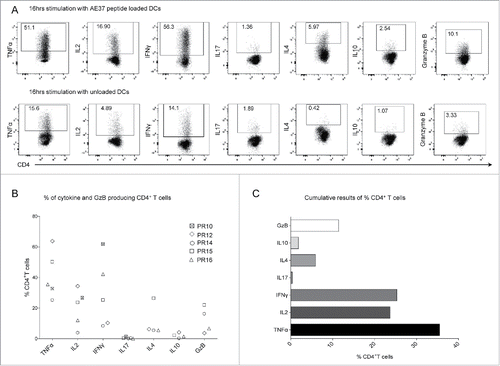
Figure 5. Multifunctional profile of AE37-responsive CD4+ T cells. (A) Pie charts show percentages of AE37-stimulated CD4+ T cells (10-d stimulation cultures) producing 1, 2, 3, or 4 cytokines among the total CD4+ T cell population. (B) Graph presents the proportions (%) of cytokine combinations that were evaluated in AE37-responsive CD4+ T cells. Multifunctional cytokine analysis was performed after stringent gating of each cytokine positive population and subsequent Boolean gating using FlowJo software.
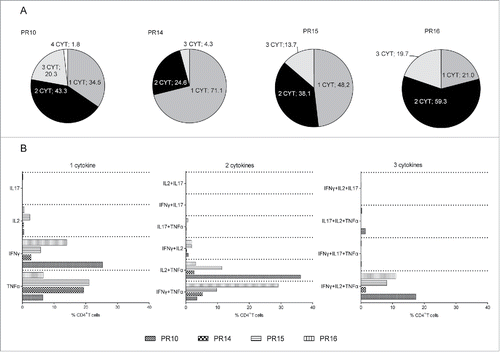
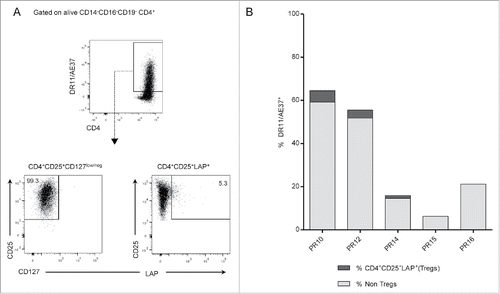
Figure 6. Assessment of Tregs in vaccine-specific CD4+ T cells. (A) Gating strategy: DR11/AE37+ cells were initially gated on live CD4+ T cells, after exclusion of CD14+CD16+CD19+ cells. Subsequent gating on tetramer+ cells, identified CD25+CD127low/neg cells and DR11/AE37+CD25+LAP+ T cells (Tregs). Dot plots from patient PR10 are presented as an example. (B) Percentages of DR11/AE37+CD4+CD25+LAP+ T cells in 10-d AE37-stimulated CD4+ T cells in all patients analyzed. Representative data at one timepoint during vaccinations with the highest Treg frequency for each patient are shown (PR10_R2, PR12_R6, PR14_R2, PR16_R2, and PR15_LTI).