Figures & data
Figure 1. Quality control of the sample scoring test. (A) Performance of scoring methods by the Kolmogorov–Smirnov (KS), T-test, and Wilcoxon tests, compared by ROC curves for the gene sets “DNA replication”, “cell division”, “cell proliferation”, and “mitosis” in the cohort of (n = 1446) B-NHL transcriptomes split according to their proliferating or non-proliferating status (determined by phenotypic analysis). (B) Dot plots of sample enrichment scores (SES) for “female” versus “male” gene sets in the (n = 553) gender-annotated samples within the dataset, using the Wilcoxon test (top), or KS-, T-test-, or Wilcoxon-based scoring (bottom). The rate of discordant cases (DC) for molecularly-inferred versus clinically-annotated gender is indicated. (C) 3D plot of the (n = 553) subtype-specified DLBCL samples using Wilcoxon-based scores for the “ABC”, “GCB”, and “PMBL” gene sets (see Fig. S2).
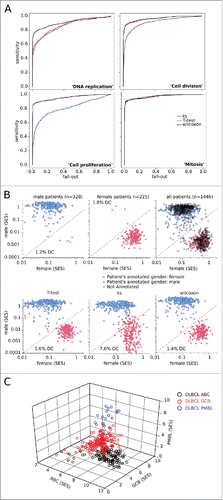
Table 1. AUC for ROC curves.
Figure 2. SES of the B-NHL data set matched to sample phenotype. Sample enrichment scores (SES) were computed for each of the 1,446 samples of the B-NHL data set and for the gene sets indicated on the y-axis. Each sample is shown by a dot, red bars are the means of the specified group. CC: centroblasts, CB: centrocytes. (A) SES for gene sets according to proliferation phenotype. (B) SES for tight junctions with sample groups split according to sample preparation. (C) SES for the KEGG gene sets “BCR signaling”, “glycolysis”, and “oxydative phosphorylation”, classified according to lymphoid malignancy.
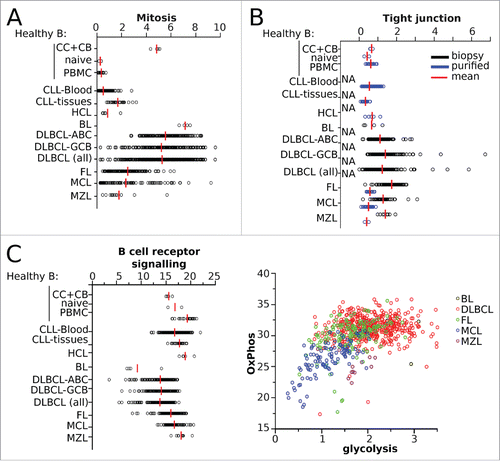
Figure 3. Scores for B-NHL tumor-associated leucocyte subsets. Distributions of sample enrichment scores (SES) for 22 leucocyte gene subsets across the B-NHL dataset, pooled by histological type of malignancy.
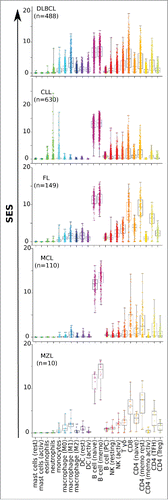
Figure 4. Scores for the immune escape gene set IEGS33 in 1,446 B-NHL samples. (A) Quality control of the IEGS33 gene set. The right skewed distribution of SES for IEGS33 in the 1446 B-NHL samples or in 500 simulated samples (shown in Fig. S2) demonstrates that this gene set carries relevant information about these samples. For B-NHL samples 24.9% of SES are above 1.3 whereas less than 5% are predicted for the simulated samples. (B–C) IEGS33 scores of the B-NHL samples classified by malignancy (B) and subtype of DLBCL (C). Red line shows group means. CC: centroblasts, CB: centrocytes.
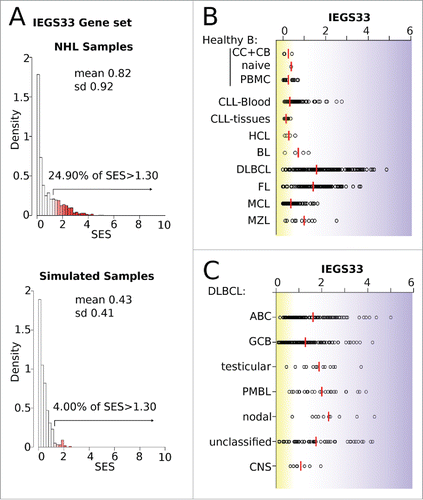
Figure 5. Stages of immune escape in B-NHL. (A) SES dot plots for IEGS33 versus T-cell activation for the 1,446 samples. CC: centroblasts, CB: centrocytes. Groups of phenotypes were arbitrarily defined according to the dotted lines: group I (IEGS33− T-cell activation−), group II (IEGS33− T-cell activation+), group III (IEGS33+ T-cell activation+), group IV (IEGS33+ T-cell activation−). (B) Left: four groups. Right: distribution of samples in these groups (group I in black, group II in green, group III in red, and group IV in blue) shown by malignancy. (C) Overall survival (OS) of the (n = 580) DLBCL patients according to the four immune escape groups (Log rank p = 0.04).
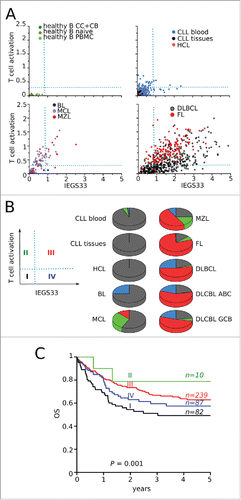
Table 2. SES in the four groups defined by T-cell activation and IEGS33.
