Figures & data
Figure 1. Treatment with PEG-rMuIL-10-induced IFNγ-dependent intratumoral IDO expression. For 1(A)–1(D), the tissue was harvested at day 24, 24 h after the last dose of PEG-rMuIL-10. (A) 4T1 tumor-bearing mice were treated for 9 d with 1 mg/kg PEG-rMuIL-10 s.c. daily and tumor growth inhibition is shown vs. control-treated mice. Closed circles represent control-treated mice, open squares represent PEG-rMuIL-10-treated mice. (B) IDO mRNA expression analysis from tumor, spleen and lymph node of mice described in (A). Open circles represent tumor tissue, open squares represent lymph node (LN), open triangles represent spleen tissue (Spl). (C) Intratumoral mRNA levels of IFNγ of mice in (A). Open circles represent control-treated mice, closed squares represent PEG-rMuIL-10-treated mice. (D) Intratumoral IFNγ protein levels of mice in (A). Open circles represent control-treated mice, closed squares represent PEG-rMuIL-10-treated mice. (E) IDO mRNA levels of in vitro cultured 4T1 tumor cells treated with PEG-rMuIL-10 alone or in combination with IFNγ. (F) Intracellular IDO1 and STAT3/pSTAT3 protein levels from AM0010 and IFNγ-treated 4T1 cells as detected by protein gel blot. IDO1 was detected after 48 h incubation with 50 ng/mL IFNγ and/or 100 ng/mL PEG-rMuIL-10. pSTAT3 and STAT3 were assessed in a time course from 0 to 20 min incubation with 100 ng/mL PEG-rMuIL-10. The protein control for pSTAT3 and STAT3 (con) was a STAT3 control cell extract purchased from Cell Signaling Technology. Statistics were determined by Students t-test where p < 0.05 is denoted by * and p < 0.01 is denoted by **.
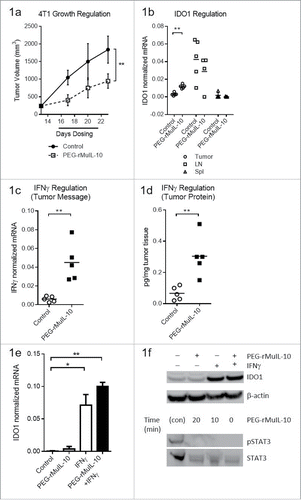
Figure 2. Treatment with an IDO inhibitor and PEG-rMuIL-10 did not exhibit combined antitumor efficacy. (A) Treatment of 4T1 tumor-bearing mice with 1mg/kg PEG-rMuIL-10 s.c. daily alters the plasma kynurenine to tryptophan ratio. Open circles represent control-treated mice, open squares represent PEG-rMuIL-10-treated mice. (B) Treatment of 42 cancer patients with AM0010 increases the serum kynurenine to tryptophan ratio. Open circles represent patients prior to treatment (Pre). Open squares represent patients after treatment (Post). (C) The cancer patient 20 μg/kg dose group serum kynurenine to tryptophan ratio. Open circles represent patients prior to treatment (Pre). Open squares represent patients after treatment (Post). (D) In vitro cell culture media IFNγ levels of activated CD8+ T cells exposed to 100 ng/mL AM0010 and 62.5, 250 or 1000 μM kynurenine for 3 d and triggered for 4 h with anti-CD3. (E) In vitro cell culture media Granzyme B levels of activated CD8+ T cells exposed to 100 ng/mL AM0010 and 62.5, 250 or 1000 μM kynurenine for 3 d and triggered for 4 h with anti-CD3. Data in (D) and (E) are representative of three donor responses. (F) Antitumor efficacy of PEG-rMuIL-10 with or without IDO inhibitor (n = 5 mice/cohort). Closed circles represent control-treated mice, open squares represent PEG-rMuIL-10-treated mice, open triangles represent IDO-inhibitor-treated mice and upside down open triangles represent PEG-rMuIL-10 and IDO inhibitor combined treated mice. Mice were treated with 1 mg/kg PEG-rMuIL-10 s.c. daily, control, IDO inhibitor (1-methyl-D-tryptophan) or the combination of the same dose of PEG-rMuIL-10 with the IDO inhibitor for 28 d. Data are representative of four total in vivo experiments. (G) Intratumoral IFNγ protein levels from two–four 4T1 tumor-bearing mice treated in (2F). (H) Intratumoral Granzyme B of the mice described in (2F). Statistics for were determined by Student's t-test where p < 0.05, p < 0.01 and p < 0.001 is denoted by *, **, ***, respectively. Statistics for was determine by use of ANOVA multiple comparisons where p < 0.05 is denoted by *.
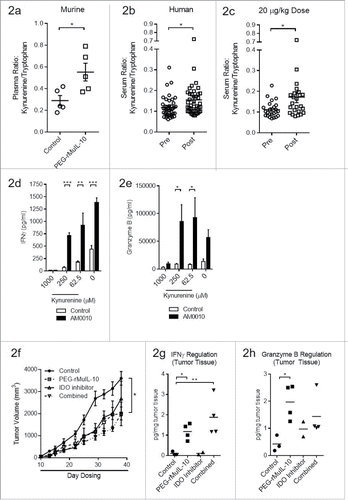
Figure 3. PEG-rMuIL-10 and AM0010 treatment decrease FoxP3+ CD4+ T cells. (A) Flow cytometric analysis of CD45+ tumor-infiltrating lymphocytes (TILs) from 4T1 tumors treated for 28 d with control, 1 mg/kg PEG-rMuIL-10 s.c. daily, IDO inhibitor (1-methyl-D-tryptophan) or the combination of the same dose of PEG-rMuIL-10 with the IDO inhibitor (n = 5 mice/cohort). Total TILs (CD45+) per gram tumor were isolated from two–four tumors per group. (B) Total CD4+ TILs from (A) per gram tumor. (C) Total CD8+ TILs from (A) per gram tumor. (D) Flow cytometric analysis of intracellular IFNγ in CD8+ TILs from (A). (E) Percentage of CD4+ or CD8+ cells within the CD45+ TIL population from (A). (F) Percentage of FoxP3+ cells within the CD4+ CD45+ TIL population from (A). Statistics for Fig. 3(E)–(F) was determine by use of ANOVA multiple comparisons, where p < 0.05 is denoted by *. Data is representative of three in vivo studies.
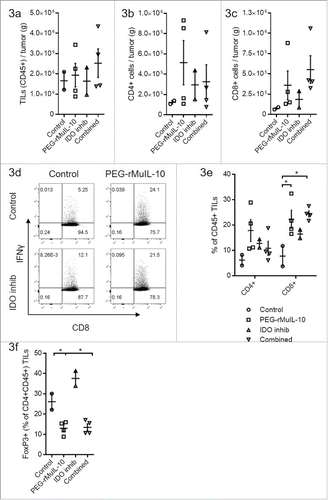
Figure 4. AM0010 exerts no effect on IDO-mediated induction of FoxP3+ CD4+ T cells in vitro. (A) IDO1 mRNA message regulation in immature or IFNγ/LPS-matured dendritic cells treated with or without 100 ng/mL AM0010 for 2 d. (B) Protein gel blot for IDO1 and STAT3/phospho-STAT3 in cells from (A). (C) Induction of FoxP3+ CD4+ T cells as per referenceCitation29 with mature dendritic cells. Human CD4+ CD25− T cells were cocultured with mature DCs for 6 d with 10 ng/mL IL-2, 100 ng/mL IFNγ and 5 µg/mL LPS with or without 100 ng/mL AM0010 and with or without 2 µM IDO inhibitor, INCB024360. T cells were then collected and analyzed by flow cytometry. Percentage indicates FoxP3+ T cells. Data is representative of four out of five donor responses.
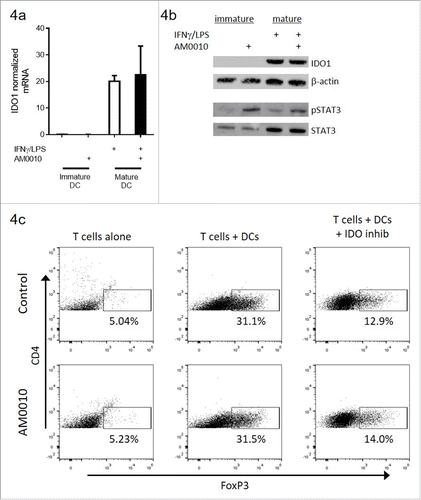
Figure 5. AM0010 treatment inhibits TGFβ/IL-2 induction of FoxP3+ CD4+ Tregs. (A) Quantitation of TGFβ plasma protein from tumor-bearing mice treated s.c. daily with 1 mg/kg PEG-rMuIL-10 or control for 10–28 d. Open circles represent control-treated mice, filled squares represent PEG-rMuIL-10-treated mice. (n = 5–8 mice/cohort) (B) Quantitation of intratumoral TGFβ protein from 4T1 tumor-bearing mice treated as in (A). Open circles represent control-treated mice, filled squares represent PEG-rMuIL-10-treated mice. (C) Assessment of human CD4+ T cells treated with 5 ng/mL TGFβ and 1000, 100 and 10 ng/mL IL-2 with or without AM0010 in vitro for 5 d. Data is representative of six of nine donor responses.
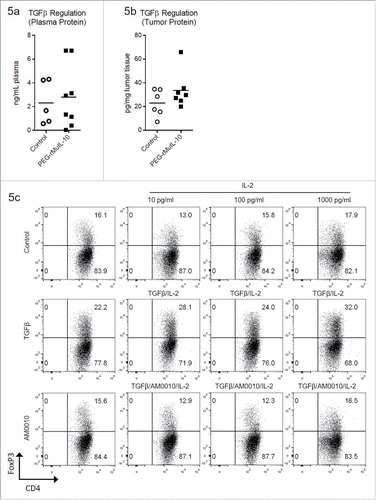
Figure 6. AM0010 does not suppress pre-polarized FoxP3+ CD4+ Tregs. The data for each plot represents the median and variation in response for each donor under the same stimulatory conditions. (A) Human CD4+ peripheral T cells were polarized for 5–7 d (with TGFβ, IL-2, anti-CD3, anti-CD28), then exposed to the stated combination of cytokines for 2–3 d and then analyzed by flow cytometry for CD4+ and FoxP3+ expression. (B) Pre-polarized cells from (A) were exposed to the identical conditions except with the addition of 2 µg/mL immobilized anti-CD3 for 2–3 d. Cells were analyzed as in (A). (C) Pre-polarized cells from (A) were exposed to the identical conditions except with the addition of both 2 µg/mL immobilized anti-CD3 and 1 µg/mL soluble anti-CD28 for 2–3 d. Cells were analyzed as in (A). Data is representative of six out of 6 donor responses. One-way ANOVA statistical analysis was performed where each treatment was compared to the No Cytokine control values for each plot. None of the differences were determined to be statistically different. In addition, each treatment group was analyzed as an independent Students t-test where the treatment group was compared to the control. In this analysis, no treatments were determined to be statistically significant.
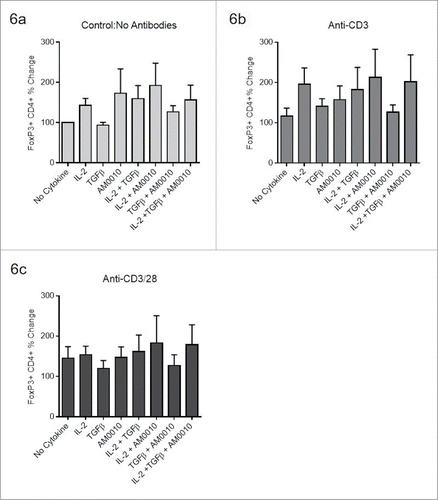
Figure 7. AM0010 treatment potentiates IFNγ+ Th1 like CD4+ T cells. (A) Quantitation of Tbet and Gata3 mRNA from tumors of mice treated s.c. daily with 1 mg/kg PEG-rMuIL-10 or control for 10–28 d. (n = 5 mice/cohort). (B) Schematic depicting in vitro experimental setup for data shown in (C–E). (C) Quantitation of secreted IFNγ, Granzyme B and Perforin from human CD4+ T cells activated for 3 d with immobilized anti-CD3/anti-CD28, then exposed for 3 d to control or 100 ng/mL AM0010. Cells were washed and then stimulated with 1 μg/mL soluble anti-CD3 for 4 h at 37°C and the proteins in the supernatant were quantified by ELISA. Data is representative of three out of three donor responses. (D) Quantitation of secreted cytokines from activated CD4+ T cells treated as in (C) with 20 ng/mL IL-12, 10 ng/mL IL-4 or 100 ng/mL AM0010. (E) Intracellular flow cytometric analysis from cells as in (D) treated with 20 ng/mL IL-12, 10 ng/mL IL-4 or 100 ng/mL AM0010. Data is representative of three out of three donor responses. Statistics in (C) were determined by Student's t-test where p < 0.05, p < 0.01 and p < 0.001 is denoted by *, **, ***, respectively. Statistics in (D) were determined by one-way ANOVA where p < 0.05, p < 0.01 and p < 0.001 is denoted by *, **, ***, respectively.
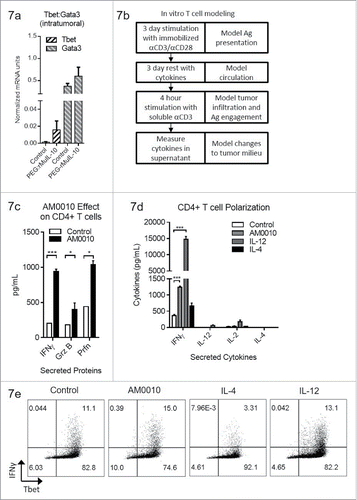
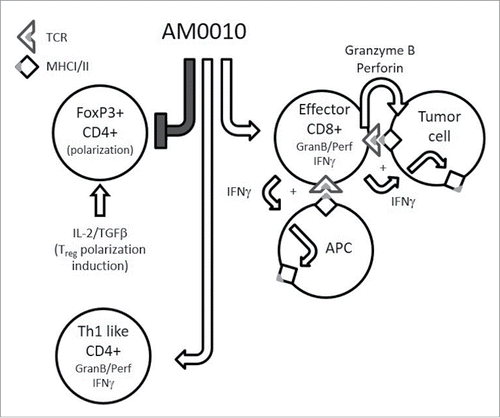
Figure 8. The pleiotropic effects of AM0010. This is a schematic cartoon of AM0010's pleiotropic effects. AM0010 treatment concomitantly and directly affects both CD4+ and CD8+ T cells. AM0010's effect on CD4+ T cells is to block IL-2/TGFβ-mediated polarization of CD4+ T cells into new FoxP3+ Tregs. This leads to a decrease in Tregs over time and decrease in TGFβ mediate immune suppression. AM0010 also facilitates the development of a Th1 IFNγ+-like CD4+ T cell. AM0010 potentiates IFNγ, granzyme and perforin production in CD8+ T cells upon TCR engagement. The potentiation of these factors then leads to a cascade of effects, not directly induced by AM0010 treatment. The IFNγ drives MHCI/II presentation on all cells in the tumor microenvironment. Therefore, both tumor cells and antigen-presenting cells express more MHCI/II+ antigen. The increased secretion of granzyme and perforin leads to more tumor cell death, increasing the antigen pool for intratumoral antigen-presenting cells to take up and present. This becomes a reiterative cycle where more tumor cell killing leads to more antigen presentation, where both immune-dominant and, we hypothesize, cryptic tumor antigens are presented. This cascade of events permits enhanced tumor immune surveillance.