Figures & data
Figure 1. Infiltration of neutrophils in HCC determines c-Met associated clinical outcome of patients. (A) Paraffin-embedded HCC samples (n = 60) were stained with anti-c-Met antibody. The representative micrographs of 60 HCC samples for c-Met staining are shown in down panel. (B) Expression of c-Met was unrelated to OS or DFS of HCC patients. 150 patients were divided into two groups according to the median value of transcriptional level of c-Met in tumor tissues. (C) 24-h production of HGF by HCC tumor-derived CD3+ T cells, CD14+ monocytes/macrophages, CD56+ NK cells, CD66b+ neutrophils, and fibroblasts (n = 4). (D) 24-h production of HGF by healthy or HCC blood-derived CD66b+ neutrophils (n = 4). (E) Paraffin-embedded HCC samples were stained with anti-CD15 antibody (n = 150). (F) Expression of c-Met was inversely correlated with OS or DFS in HCC patients with high peritumoral neutrophil infiltration. The patients were divided into four groups according to the median value of c-Met expression and peritumoral neutrophil infiltration in HCC tissues. Scale bar, 100 µm. Results represent mean ± SEM. *p < 0.01.
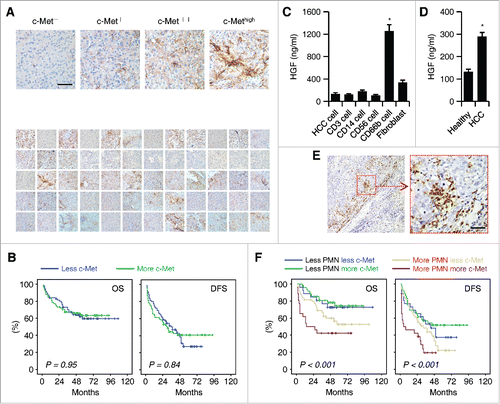
Table 1. Clinical characteristics of the 225 HCC patients.
Table 2. Univariate and multivariate analyses of factors associated with survival and recurrence.
Figure 2. Activation of Erk1/2, p38, and NF-κB is essential for the induction of HGF in tumor associated neutrophils. (A–C) Purified neutrophils were left untreated or stimulated with culture supernatant from primary HCC cells, three heptatoma tumor cell lines or a normal liver cell line (L02) for 48 h. (A) The release of HGF in the culture supernatants at the indicated times were determined by ELISA. (B) The expression of HGF in neutrophils after 2-h treatment was detected by real-time PCR. (C) The activation of Erk1/2, p38, JNK, AKT, and IκBα at 0.5 h was measured by immunoblotting. (D) Inhibition of Erk1/2, p38, and IκBα impaired HGF expression in neutrophils exposed for 24 h to culture supernatant from primary HCC cells. The illustrated results represent mean ± SEM of four separate experiments. *p < 0.05, **p < 0.01, compared with medium; #p < 0.05, compared with DMSO.
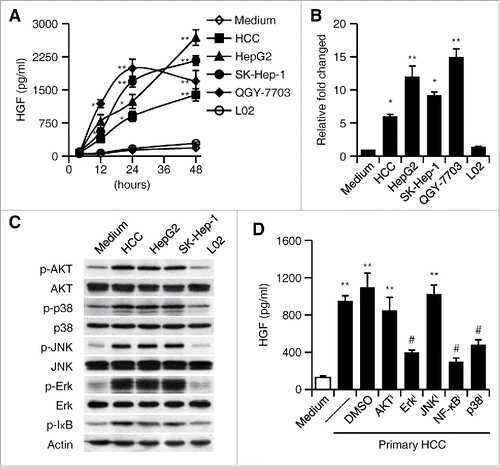
Figure 3. GM-CSF is responsible for the induction of HGF in tumor neutrophils. (A) Plasma concentration of GM-CSF in healthy donors (n = 22) and HCC patients (n = 39 for stage I+II and n = 36 for stage III+IV). Horizontal bars represent median values. (B) The concentration of GM-CSF in the culture supernatants from primary or established hepatoma cells. (C) Effect of GM-CSF on neutrophil HGF production after 24-h stimulation at indicated concentration (n = 4). (D) Inhibition of Erk1/2, p38, and IκBα impaired GM-CSF-mediated HGF production in neutrophils (n = 4). (E) Neutralization of GM-CSF attenuated the HGF production in neutrophils treated with culture supernatants from hepatoma cells (n = 4). (F) shGM-CSF in Hepa1-6 cells attenuated the GM-CSF expression in hepatoma tissues from mice and the HGF production in neutrophils derived from hepatoma tissues (n = 5 for each group). Results represent mean ± SEM. *p < 0.01, compared with those treated with DMSO, IgG, or shNC in corresponding group.
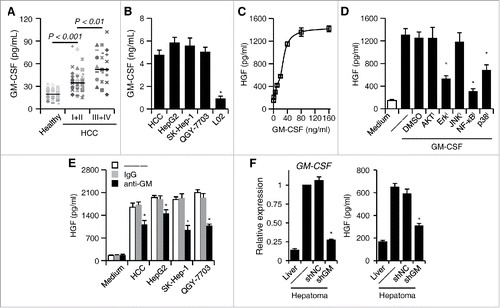
Figure 4. The effect of HGF/c-Met axis on the migration and invasiveness of tumor cells. (A and B) Effects of TCM or BCM on hepatoma QGY-7703 cell migration (A) and invasion (B); untreated QGY-7703 cells, Med. (C) Suppression of c-Met activity by inhibitor SU11274 (left panel) or neutralization of HGF in TCM (right panel) effectively impaired hepatoma QGY-7703 cell migration. (D) Suppression of c-Met activity by inhibitor SU11274 effectively impaired hepatoma QGY-7703 cell migration (left panel) and invasion (right panel) stimulated by CM from GM-CSF-treated neutrophils. (E) Suppression of Erk1/2, p38, and IκBα activities in neutrophils treated with GM-CSF attenuated the effects of neutrophil CM on hepatoma QGY-7703 cell migration and invasion. All the data shown are representative of at least four separate experiments. Scale bar, 100 μm. *p < 0.01, compared with medium in D,E; #p < 0.05, compared with DMSO.
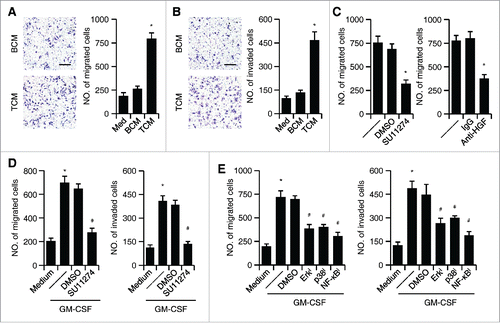
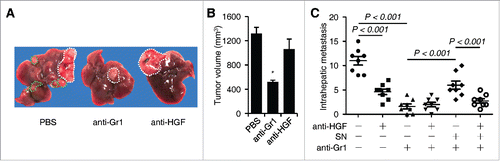
Figure 5. Effects of neutrophils and HGF on hepatoma growth and intrahepatic metastasis in tumor-bearing mice (n = 8 for each group). (A–C) The Hepa1-6 hepatoma were treated with PBS or injected with anti-Gr1 and/or anti-HGF in the presence or absence of culture supernatant (SN) from coculture of blood neutrophils and Hepa1-6 cells as described in the methods. The representative images hepatoma with intrahepatic metastasis was shown in (A). The numbers of intrahepatic metastasis in different group were determined by immunohistochemical staining of hematoxylin and eosin (C). n = 8 for each group; *p< 0.01.