Figures & data
Figure 1. Identification of PTX3 enhancers by histone modification analysis. (A) Analysis of H3K4me1, H3K4me3, H3K9Ac, H3K27Ac, and H3K27me3 histone modifications at enhancer 1, promoter and enhancer 2 by ChIP in human macrophages in basal and inflammatory conditions (TNFα 20 ng/mL, 4h). Results are expressed as fold change relative to IgG (N = 3 experiments). *p ≤ 0.05; Student's t-test. (B) Analysis by RT-qPCR of PTX3 mRNA expression in HCoEpic and 8387 cells and in human macrophages in basal condition and after TNFα treatment (20 ng/mL, 4h). PTX3 mRNA expression is expressed as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01; Student's t-test. (C) Correlation analysis between the enrichment of H3K27Ac on enhancer 1 and the expression of PTX3 mRNA in HCoEpic and 8387 cells and in macrophages. N = 4 experiments for macrophages and 8387 cells and N = 2 for normal human colonic cells.
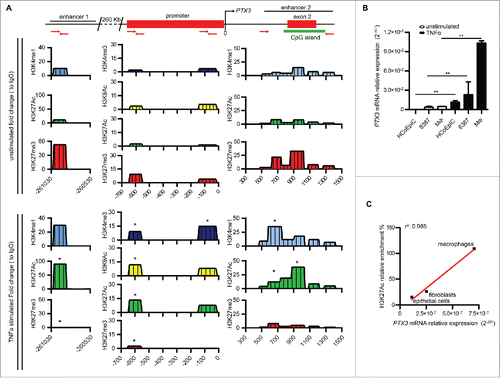
Figure 2. Analysis of inflammatory TFs and transcription preinitiation complexes regulating PTX3 enhancer activity. (A) ChIP assay for inflammatory TFs [NF-κB (p65), c-Fos, c-Jun, Sp1, and Pu.1] in macrophages (top panel) and 8387 cells (bottom panel) in basal and inflammatory (TNFα 20 ng/mL, 4 h) conditions. (B) ChIP assay for TAF1 and RNA Pol II in macrophages in basal and inflammatory conditions. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student's t-test.
![Figure 2. Analysis of inflammatory TFs and transcription preinitiation complexes regulating PTX3 enhancer activity. (A) ChIP assay for inflammatory TFs [NF-κB (p65), c-Fos, c-Jun, Sp1, and Pu.1] in macrophages (top panel) and 8387 cells (bottom panel) in basal and inflammatory (TNFα 20 ng/mL, 4 h) conditions. (B) ChIP assay for TAF1 and RNA Pol II in macrophages in basal and inflammatory conditions. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student's t-test.](/cms/asset/9a7036df-1a2f-4581-a9c9-59457a9bc1b7/koni_a_1333215_f0002_oc.gif)
Figure 3. PRC2 complex regulates RNA Polymerase II binding on PTX3 regulatory regions. (A) ChIP assay for EZH2 and Suz12 in macrophages in basal and inflammatory (TNFα 20 ng/mL, 4 h) conditions. (B) ChIP assay for H3K27me3, RNA Pol II and TAF1 in 8387 cells in basal condition and after stimulation with GSK343 1 μM for 48 h. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01; Student's t-test.
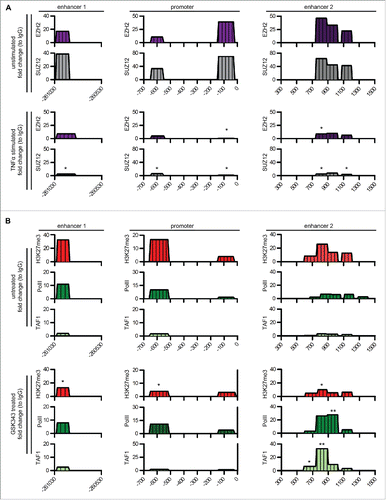
Figure 4. PTX3 epigenetic modifications in human cancers. (A, B) Analysis by Methylated CpG Island Recovery Assay (MIRA) of the percentage of methylation enrichment of PTX3 regulatory regions in human mesenchymal and epithelial tumors (A) and CRC (B) and in their healthy counterparts. Results are expressed as percentage of enrichment relative to input DNA normalized on a positive control and represented as mean ± SEM. (N = 6 samples for human healthy colon, N = 3 samples for low-grade colon adenoma, N = 5 samples for high-grade adenoma, N = 2 for human healthy counterpart, N = 12–14 for CRC tumor stages). *p <0.05, **p <0.01, Student's t-test. (C) Analysis of H3K4me1, H3K27Ac, and H3K27me3 histone modifications by ChIP in human CRC stages I, II, and III. Results are expressed as fold change relative to IgG and as mean ± SEM. (N = 3 experiments). *p ≤ 0.05, **p ≤ 0.01; Student's t-test. (A–C) Regions analyzed are reported in the upper part of the panels.
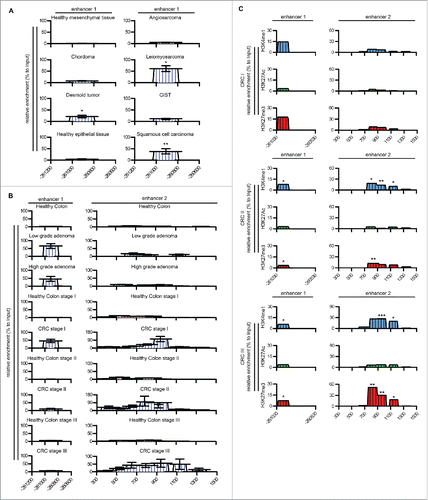
Figure 5. Effect of methylation inhibition on PTX3 mRNA expression and enhancer activity. (A) PTX3 mRNA expression by RKO (left panel) and HCT116 (right panel) cells upon treatment with TSA (150 nM for 48 h) or 5-AZA-dC (15 μM for 72 h) and TNFα (20 ng/mL). Results are expressed as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA. (B) ChIP assay for H3K4me1, H3K4me3, H3K9Ac, H3K27Ac, H3K27me3 in RKO cells upon treatment with TNFα and 5′AZA-dC. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean (N = 2 experiments). *,ˆ,$p ≤ 0.05; **,ˆˆ,$$p ≤ 0.01; ***p <0.001. Student's t-test. ˆ: unstimulated vs. 5′AZA-dC; $: 5′AZA-dc vs. 5′AZA-dC + TNFα; *TNFα vs. 5′AZA-dC + TNFα.
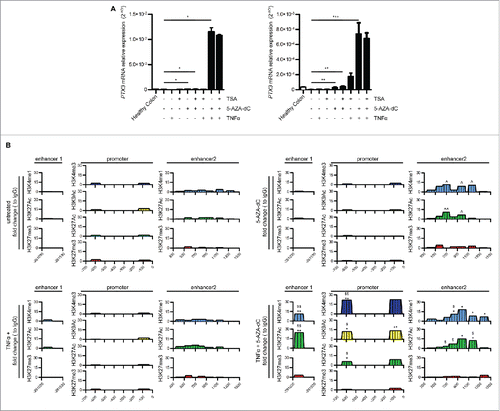
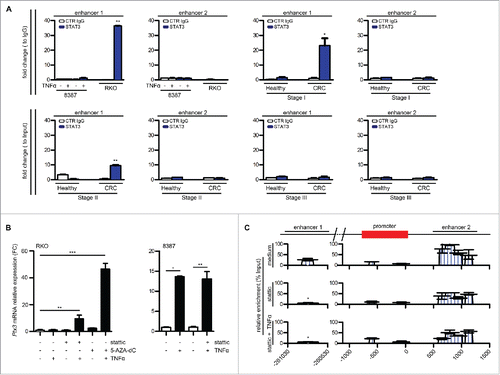
Figure 6. Role of STAT3 in PTX3 epigenetic modification. (A) ChIP assay for STAT3 in RKO and 8387 cells in basal condition and in CRC stage I, II, and III and in their normal counterparts. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean ± SEM (N = 2 experiments for the cell lines and CRC stage II, N = 2 healthy colon samples for each tumor stage and N = 4 for CRC stage I and II). *p ≤ 0.05, **p ≤ 0.01, Student's t-test. (B) PTX3 mRNA expression in RKO and 8387 cells treated with 5-AZA-dC 15 μM for 72 h, Stattic 1 μM for 48 h, and TNFα 20 ng/mL for 4 h, as specified. Results are expressed as mean ± SEM and as fold change of basal conditions (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA (left panel) or Student's t-test (right panel). (C) Analysis by MIRA of the percentage of methylation enrichment of PTX3 regulatory regions in RKO cells after treatment with stattic and TNFα as specified. Results are expressed as percentage of enrichment relative to input DNA normalized on a positive control and represented as mean ± SEM. (N = 2 experiments) *p < 0.05. Student's t-test.