Figures & data
Table 1. Characteristics of AML patient.
Figure 1. Heterogeneity of cytotoxic potential and surface receptor expression among evNK issued from different UCB-donors. (A) Cytotoxic activity of evNK issued from 4 different donors against K562 target cells. Cytotoxicity was determined by a conventional 4-hr Cr51-release assay at different E:T ratios(100:1, 30:1, 10:1, 3:1).(B) Flow cytometry analysis of the surface expression of the indicated NK receptor on evNK derived from UCB-donors of . Percentage of positive cells was measured on CD56-purified evNK cells.
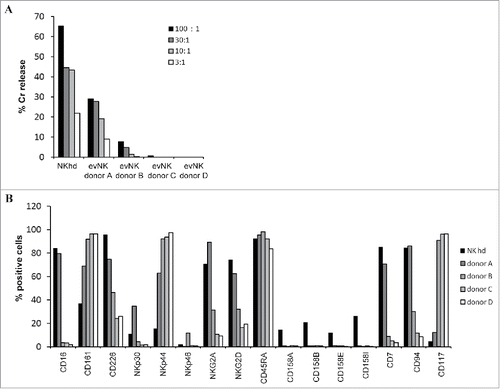
Figure 2. CD94 surface expression on evNK correlates with higher lytic potential. Cytotoxic activity of evNK derived from different UCB-donors was measured against K562 (A), U937 (B), and HL-60 (C) target cells at different E:T ratios (30:1, 10:1, 3:1, 1:1) by a conventional 4-hr Cr51-release assay. EvNK were either sorted based on CD94 surface expression (evNK CD94+; evNK CD94−) or not (evNK).
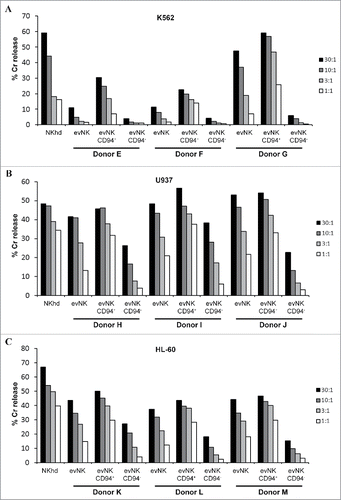
Figure 3. CD94-positive evNK display higher expression of NKG2 receptors and LFA-1. (A) (B) (C) Comparative flow cytometry analysis of surface expression ofthe indicated NK receptors on evNK derived from 3 UCB donors and sorted (CD94+; CD94−) or not (Tot) based on CD94 surface expression. Percentage of positive cells was measured on CD56-purified evNK cells (Tot), CD56-purified then CD94-positive sorted evNK cells (CD94+), and CD56-purified then CD94-negative sorted evNK cells (CD94−).
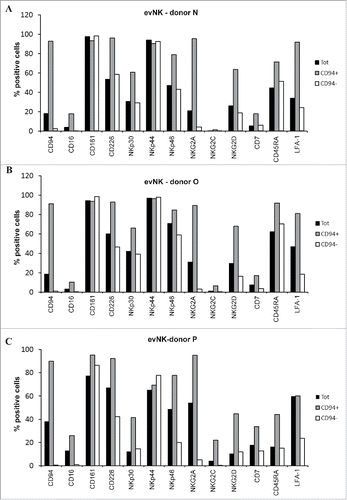
Figure 4. CD94-positive evNK are efficient to form immune synapses with target leukemic cells unlike CD94-negative ones. (A) Quantification of CD94-positive and CD94-negative evNK involved in immune synapse formation with K562, U937, and HL-60target cells. EvNK were sorted based on CD94 expression and CD94-positive and –negative evNK were subsequently co-cultured for 30 min with target leukemic cells. Results are means of 2 independent experiments and are expressed as percent of evNK cells involved in immune synapses/total evNK. *p < 0,05. (B) Representative illustration of immune synapse formation between K562, U937, or HL-60 leukemic cells and CD94+-evNK. Immune synapses are visualized by F-actin staining with rhodamine-phalloidine (purple). DAPI (blue) is used for nuclei labeling. Scale bars, 5 µm. L: leukemic cells; NK: NK cells.
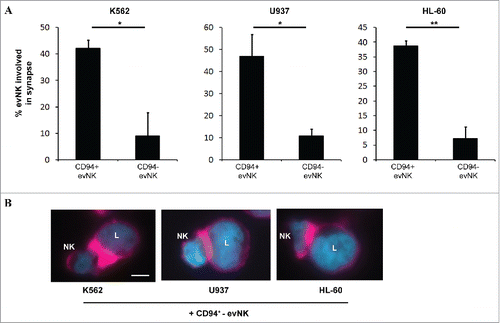
Figure 5. Patient-derived AML cells are resistant to evNK and NKhd. Cytotoxic activity of evNK and NKhd was measured against patient-derived AML cells at different E:T ratios (30:1, 10:1, 3:1, 1:1). K562 cells were used as positive control of NK cytotoxicity.
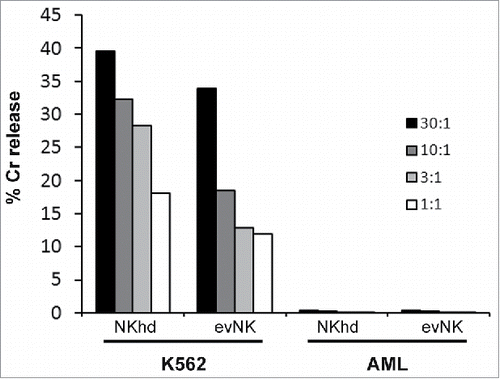
Figure 6. Organ distribution of evNK and NKhd in AML-engrafted mice. 5.106 evNK or NKhd were intravenously injected per mouse each 3–4 d for 14 d in AML-engrafted NSG mice. Percentages (A) and absolute numbers (B) of evNK and NKhd isolated from blood, spleen, and BM of AML-engrafted mice. EvNK and NKhd were identified using anti-human CD45 and CD56 antibodies. Mouse cells were excluded using anti-mouse CD45 antibody. n = 6–7 for evNK or NKhd groups.
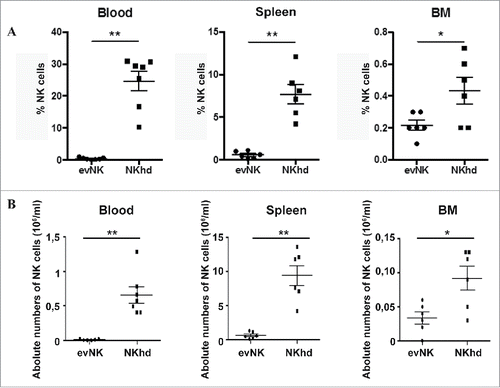
Figure 7. Absence of anti-leukemic effect of evNKand NKhd in AML-engrafted mice. Percentages (A) and absolute numbers (B) of AML cells isolated from blood, spleen, and BM of AML-engrafted NSG mice at day 14 after evNK and NKhd intravenous injection. AML were identified using anti-human CD45 and CD34 antibodies. Mouse cells were excluded using anti-mouse CD45 antibody. n = 6–7 for control, evNK, or NKhd groups.
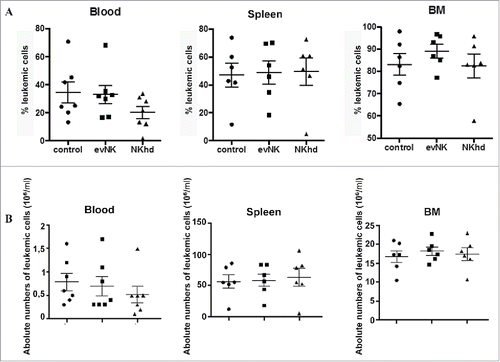
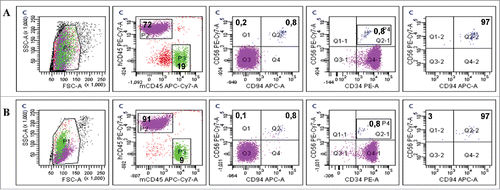
Figure 8. CD94-positive evNK interact with AML cells. Representative dot plots of CD56, CD94, and CD34 expression in blood isolated from AML-engrafted NSG mice injected with either evNK (A) or NKhd (B) at day 14 after NK cell intravenous injection. Untreated AML-engrafted mice were used as control (C). Anti-human CD45, CD56 and CD94 antibodies were used for NK cell identification. Anti-human CD45 and CD34 antibodies were used for AML cell identification. Bold numbers represent the percentage of cells in the corresponding quadrant. The percentages indicated in the third to fifth columns are calculated within human cells (P2 gate, hCD45+).
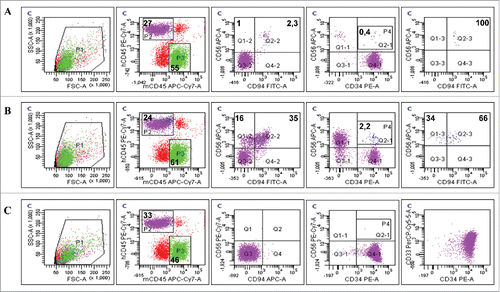
Figure 9. Absence of delayed anti-leukemic effect by evNK of NKhd at day 28 post-NK injection. Absolute numbers of AML (A) and evNK/NKhd (B) cells in the blood, spleen, and BM of AML-engrafted NSG mice at day 28 after the first NK intravenous injection. n = 1 for control, evNK, and NKhd groups.
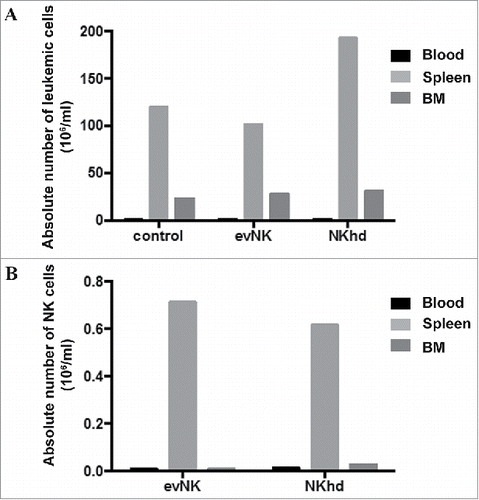
Figure 10. Residual evNK at day 28 are CD94-positive. Representative dot plots of CD56, CD94 and CD34 expression in blood isolated from AML-engrafted NSG mice injected with either evNK (A) or NKhd (B) at day 28 after the first NK cell intravenous injection. Anti-human CD45, CD56 and CD94 antibodies were used for NK identification. Anti-human CD45 and CD34 antibodies were used for AML identification. Bold numbers represent the percentage of cells in the corresponding quadrant. The percentages indicated in the third to fifth columns are calculated within human cells (P2 gate, hCD45+).