Figures & data
Figure 1. Frequency of infiltrating and circulating γδ T cells expressing either Vδ1 or Vδ2 TCR δ chains in HD and CRC patients. (A) Cumulative analysis of immune infiltrates of 70 colon cancer specimens. Lymphomonocyte populations were evaluated by the use of cell-surface markers and indicated as percentage of the total number of CD45+ cells in each sample. (B) Box plot of percentages of Vδ1 or Vδ2 γδ T cells subsets in healthy tissue, tumor tissue and peripheral blood of CRC patients and peripheral blood of HD subjects. Boxes represent 25th to 75th percentiles; middle bar identifies median; whiskers show minimum and maximum. *p<0.05 performed by nonparametric Mann-Whitney test, unpaired and 2-tailed with confidential interval 95%. (C) Representative dot plots of the gating strategy used to define Vδ1 and Vδ2 T cells from healthy and tumor tissues. The following gating strategy was used to detect γδ T lymphocytes: FSC/SSC, single cells, live cells CD45/CD3, Vδ1 and Vδ2 T cells. (D) Sections from CRC patients were stained with anti-human pan-γδ TCR (red) and anti-CD3 (green) for immunofluorescent (IF) staining. Right panel is a magnified view and the arrows display the colocalization of γδ TCR and CD3. Nuclei were contrasted with DAPI. One of 3 independent experiments is shown. (E) Phenotypical analysis of Vδ1 and Vδ2 T cells among healthy and tumor tissues and PBMC of CRC patients, upon staining with mAbs to CD45RA and CD27, and gating on CD3+ Vδ1+ or CD3+ Vδ2+ T cells. Beside, flow cytometry panels of a representative dot plot. Isotype-matched mAbs were used as controls. Viable lymphocytes were gated by forward and side scatter, and analysis was performed on 100,000 acquired events by using FlowJo. PBMC were stained with anti-CD3, anti-Vδ2, anti-CD45RA and CD27 mAbs.
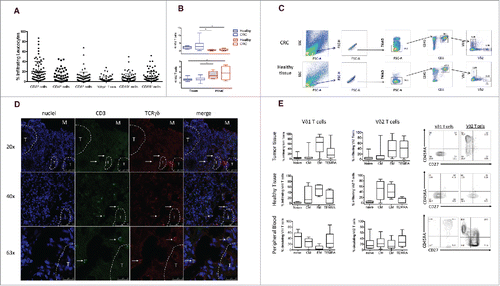
Figure 2. Cytokine production of tumor infiltrating γδ T cells. (A) Box plots of cumulative data of healthy tissue and tumor tissue samples from 20 CRC patients. Cells were stimulated in vitro as described in Materials and Methods and were stained with mAbs to IFN-γ, IL-17 and TNF-α. *p<0.05 and **p<0.01 performed by nonparametric Mann-Whitney test, unpaired and 2-tailed with confidential interval 95%. (B) Flow cytometry analysis of healthy and tumor tissue from one representative CRC patient. (C) Representative dot plots to define IL-17 or IFN-γ producing γδ, Vδ1 and Vδ2 T cells gated separately on CD45+ CD3+γδ−, CD45+ CD3+ Vδ1+ or CD45+ CD3+ Vδ2+ T cells. (D) Representative dot plots to define cells making IL-17 or IFN-γ upon gating on CD45+ IL-17+ or CD45+ IFN-γ+ cells, of healthy and tumor tissue. (E) Pearson correlation of TCR, IFNG and IL17A gene expression levels in n = 585 CRC tumor samples.**p<0.01. (F) CRC-infiltrating CD45+ single cells were used to generate the SPADE tree, and were grouped in 2 different populations, CD3− and CD3+(black outer circles). The distribution of the major populations is showed for one representative sample. The branching tree is based on the number of cells included in each node and the legend indicates the range of cell per node according to relative median fluorescence intensity.
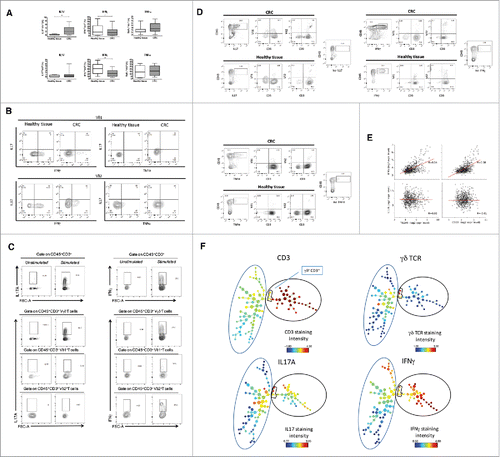
Figure 3. CSC supernatants inhibit proliferation and IFN-γproduction by γδ, CD4 and CD8 T cells. (A) Cumulative data (n = 5) from proliferation assay. Positive control (CTRL+) and negative control (CTRL-) refer to cells stimulated with PHA and unstimulated cells, respectively. Data are mean percentage of positive cells ± SD. Shown also histogram plots of proliferation of γδ T cells upon culture with PHA and in the presence of CAF and CSC supernatants. (B) Frequency of IL-17- or IFN-γ-producing γδ T cells upon incubation for 24 or 48 hrs with PHA in the presence of CAF or CSC supernatant. Histograms show cumulative data from 5 different experiments. Error bars indicate SD. Shown are also representative dot plots.(C) Cumulative and flow cytometry analysis of IFN-γ and IL-17 production by CD4 and CD8 T cells upon incubation for 48 hrs with PHA in the presence of CAF or CSC supernatant.*p<0.05.
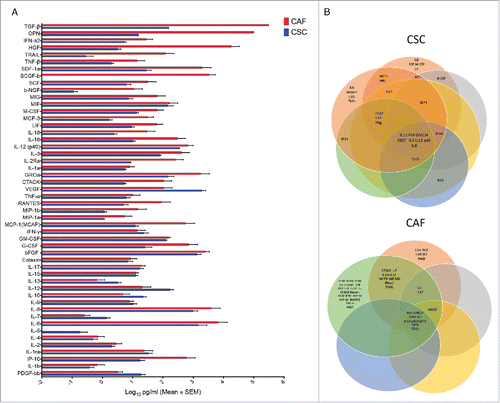
Figure 4. Comparative analysis of 50 different cytokines in CSC and CAF secretome (A) Levels of 50 different cytokines in 48 hrs supernatants of CSC and CAF by the Luminex platform. (B) Cytokine grouping in 6 CRC and 5 CAF samples as represented by Venn diagrams.