Figures & data
Figure 1. Life-cycle models of eosinophils. Eosinophils are produced in the bone marrow through a series of progenitors from the most immature stem cells (Hematopoietic Stem Cells: HSC), Multipotent Progenitor Cell: MPP) to committed eosinophil progenitors (Semper, Muehlberg et al.) that finally generate mature eosinophils. This process of proliferation and differentiation is driven by transcription factors (e.g. GATA-1). IL-5, together with IL-3 and GM-CSF, controls eosinophil development in the bone marrow. Recent evidence indicates that IL-33 precedes IL-5 in regulating eosinophilopoiesis via the activation of the IL-33R receptor, ST2. Circulating human eosinophils selectively express IL-5Rα, CCR3, EMR1, CRTH2, and Siglec-8. Eosinophil can leave the bloodstream and target the bone marrow, liver, and spleen for elimination. In some tissues, eosinophils can be phagocytosed by macrophages. Eosinophils express a wide spectrum of integrins (α and β) and can roll and adhere to VCAM-1 and ICAM-1 on activated endothelial cells. This interaction favors the chemotactic activity of several chemokines (eotaxin-1/CCL11, eotaxin-2/CCL24, eotaxin-3/CCL26, and RANTES) that activate the CCR3 receptor highly expressed on eosinophils. This explains the propensity of eosinophils to leave the bloodstream and migrate into inflamed tissues and certain tumors. Several cells (fibroblasts, epithelial and endothelial cells, smooth muscle cells, T cells, macrophages, and eosinophils itself) are a major source of these chemokines. IL-5, IL-33 and presumably other cytokines locally produced by both immune cells (eosinophils, ILC2, T cells, and mast cells) in tumor microenvironment and by tumor cells can prolong the life span of eosinophils at site of tumor growth.
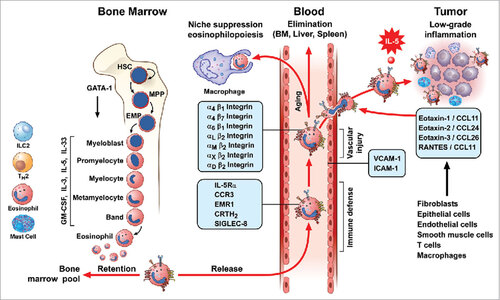
Table 1. Anti-tumorigenic activity of eosinophils in experimental tumors.
Table 2. Role of eosinophils in clinical cancers.
Figure 2. Role of eosinophils in survival rate of cancer patients. Kaplan-Meier plots depict the overall survival rate of all the indicated cancer types in function of the expression of indicated genes in cancer tissues. Survival events (vertical black lines) of patients were stratified based on the low (red color) or high expression (green color) of Siglec8 (Sialic Acid Binding Immunoglobulin-like Lectin 8, GenBank/Entrez ID: 27181), EPX (Eosinophil Peroxidase, GenBank/Entrez ID: 8288), and CLC (Charcot-Leyden crystal protein, GenBank/Entrez ID: 1178), three specific markers for human eosinophils. The survival rate data were extracted from patients in publicly available expression array datasets by the web-based tool SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). Each graph depicts a distinct patient's cohort. LS, Liposarcoma; DLBCL, Diffuse Large B cell lymphoma; AC, Adenocarcinoma; UC: Urothelial carcinoma; GB, Glioblastoma. PCox, Log-Rank (Mentel-Cox) P value calculated by SurvExpress. P values below 0.05 were considered statistically significant.
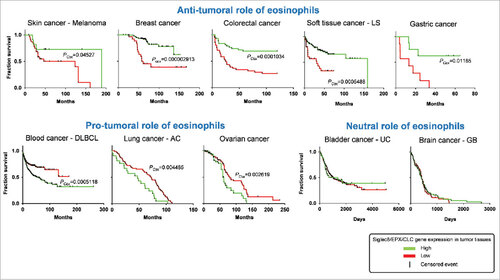
Figure 3. Roles of eosinophils in human and experimental tumors. In green boxes are indicated those tumors in which eosinophils play a protective role. In red boxes those tumors in which eosinophils play a pro-tumorigenic role. In mixed red/green boxes are presented tumors in which eosinophils play both a pro- and anti-tumorigenic role in different studies.
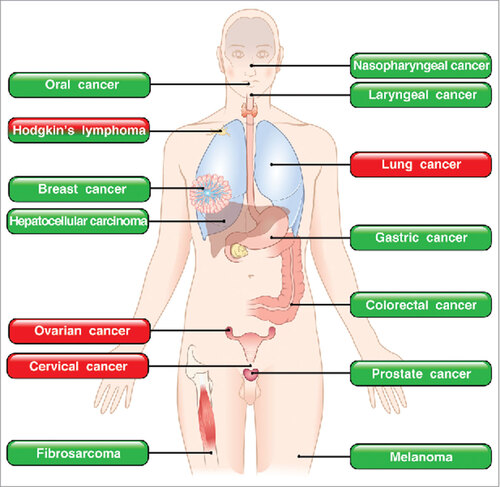
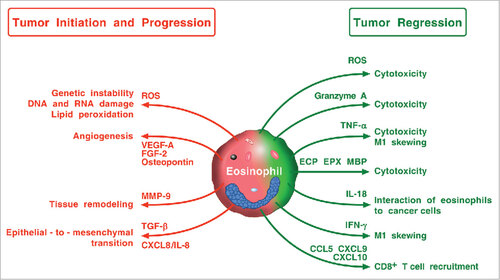
Figure 4. Possible mechanisms by which eosinophils and their mediators play a protumorigenic or an antitumorigenic role. Eosinophils in TME can promote tumor initiation and progression through the release of ROS, angiogenic factors, metalloproteinase-9, and the induction of epithelial-to-mesenchymal transition. Eosinophils can exhibit antitumor activity through direct tumor cell cytotoxicity mediated by ROS, granzyme, TNF-α, eosinophil cationic proteins (ECP, EPX, MBP), and IL-18. IFN-γ produced by eosinophils favors M1 polarization of TAMs. Eosinophils can also exert antitumor activity indirectly through the attraction of CD8+ T cells via the production of CCL5, CXCL9, and CXCL10.