Figures & data
Figure 1. Priming and expansion of antigen-specific pmel-1 CD8+ T cells by cancer vaccines. To increase the frequency of naive gp100-specific CD8+ T cells, 1 × 107 spleen cells (14.5 ± 2.7% of which were CD8+Thy1.1+hgp100 tetramer+) from pmel-1 transgenic mice were adoptively transferred into C57BL/6 mice prior to vaccination. These mice then received SP-V or DC-V. Vaccines were administered on days 0 and 14. On day 28, spleen cells from vaccinated mice were analyzed by flow cytometry to evaluate the quantity and quality of vaccine-induced pmel-1 CD8+ T cells. (a) Pmel-1 CD8+ T cells were detected as CD8+CD90.1+ cells by flow cytometry. One plot from each group is depicted. Numbers indicate the frequency of CD8+CD90.1+ cells from a representative mouse. Percentages (b) and absolute cell numbers (c) of CD8+CD90.1+ cells are shown. (d) CD44 and CD62L expression by CD8+CD90.1+ cells. Numbers indicate the frequency for each quadrant from a representative mouse. Bar graphs demonstrate percentages of pmel-1 cells with the CD44hiCD62L+ central memory phenotype (e) and the CD44hiCD62L− effector memory phenotype (f). (g) CD127 and CD62L expression by CD8+CD90.1+ cells. (h) Percentages of CD127+ cells. (i) PD-1 and Tim-3 expression by CD8+CD90.1+ cells. Bar graphs display percentages of PD-1+ (j) Tim-3+ (k) and PD-1+Tim-3+ (l) cells. Data are representative of three experiments with 4 mice per group.
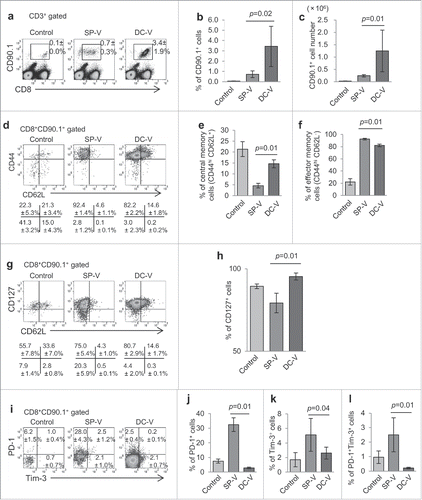
Figure 2. Proliferative capacity and effector function of vaccine-induced pmel-1 cells. C57BL/6 mice received vaccination as described in the legend to . On day 28, spleen cells were labeled with CFSE and cultured in the presence or absence of hgp100 peptide at 1 µg/ml for 2 days. (a) CFSE dilution in CD8+CD90.1+ cells was determined by flow cytometry. One of the plots from each group is depicted. Numbers indicate the percentage of cells which had divided more than three times. (b) Bar graphs indicate the percentages of cells dividing more than 3 times. Data are representative of two experiments with 5 mice per group. IFNγ (c) and TNFα (d) production and CD107a translocation (e) of CD8+CD90.1+ pmel-1 cells stimulated with or without hgp100 peptide. One plot from each group is depicted. Numbers indicate the frequency of IFNγ+ cells from a representative mouse. Frequencies (f, g, h) and absolute cell numbers (i, j, k) of IFNγ+ (f, i), TNFα+ (g, j) and CD107a+ (h, k) cells. Data are representative of three experiments with 4 or 5 mice per group.
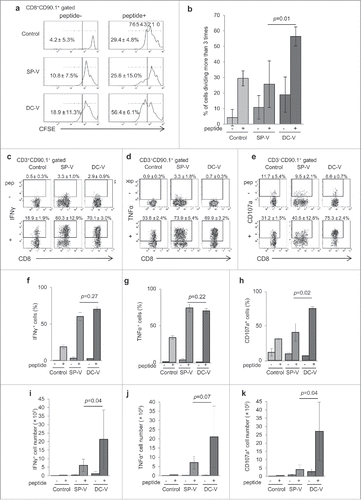
Figure 3. Gene expression profiles of naïve, SP-V-primed and DC-V-primed pmel-1 cells. Mice were treated as described in the legend to . CD8+CD90.1+ cells were sorted from pooled spleen cells of the indicated groups of mice (19 SP-V and 10 DC-V mice). Gene expression was analyzed by microarray. (a) Heat map of hierarchical clustering analysis based on fold-changes of gene expression relative to mean values. Genes of interest were categorized and show (b) inhibitory receptors, (c) cytokines, (d) M phase, (e) S phase and DNA replication, (f) glycolysis, gluconeogenesis and TCA cycle, (g) amino acid metabolism, (h) fatty acid metabolism, (i) electron transport chain in mitochondria.
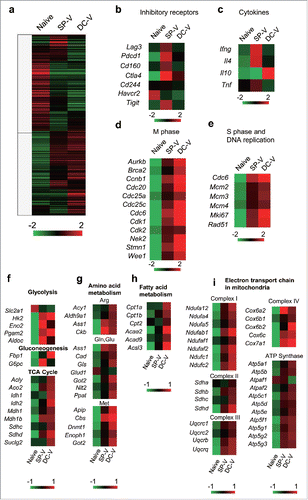
Figure 4. Glucose uptake and mitochondrial membrane potential of vaccine-primed pmel-1 cells. Mice were treated as described in the legend to . (a) Uptake of 2-NBDG in CD8+CD90.1+ cells was determined by flow cytometry. Mitochondrial mass (b) and membrane potential (c) in CD8+CD90.1+ pmel-1 cells evaluated by labeling with Mitotracker Green (b) and TMRE (c) staining. One plot from each group is depicted. Bar graphs indicate mean fluorescence intensities of 2-NBDG (d) and frequencies of Mitotracker Greenlow (e) and TMRElow (f) cells in CD8+CD90.1+ pmel-1 cells. Data are representative of two experiments with 5 mice per group.
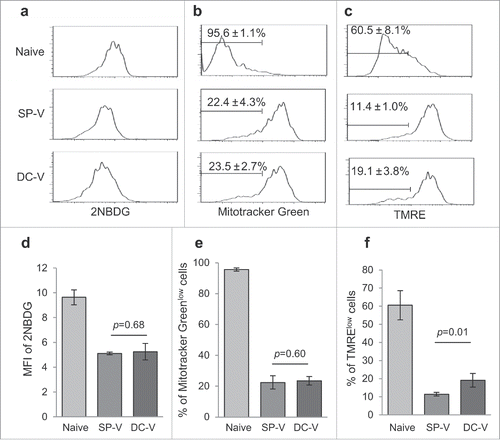
Figure 5. In vivo anti-tumor activity of SP-V and DC-V. (a) Mice (5 mice per group) were treated as described in the legend to . Two weeks later, they were challenged by subcutaneous injection of 1 × 106 B16F10 cells and tumor growth was monitored. The graphs show tumor volume of individual mice. The tumor volumes were compared on day 17. Data are representative of two experiments with 5 mice per group. (b) For the therapeutic model, C57BL/6 mice (6 mice per group) were first inoculated with 1 × 106 B16F10 cells subcutaneously on day 0. On day 5, mice received 1 × 107 naïve pmel-1 transgenic spleen cells. SP-V or DC-V was then administered twice on days 5 and 12 to these animals and tumor growth monitored. The graphs show tumor volume of individual mice. The tumor volumes were compared on day 18. Data are representative of three experiments.
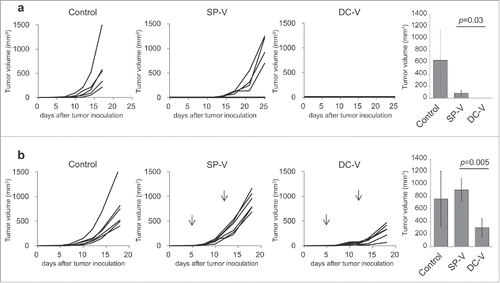
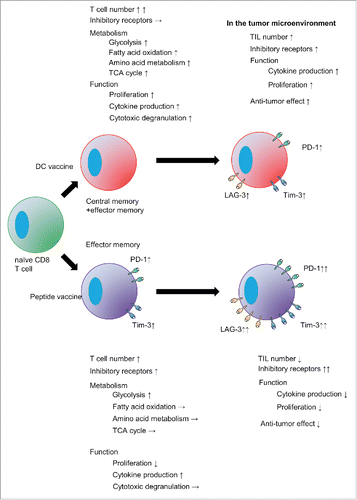
Figure 6. Phenotype and function of vaccine-primed pmel-1 cells in the tumor microenvironment. Mice were treated as described in the legend to . On day 18, tumor-infiltrating cells were isolated. Tumor-infiltrating leukocytes (a) and pmel-1 cells (b) were detected as CD45+ and CD45+CD8+CD90.1+ cells, respectively. One plot from each group is depicted. Frequencies (left) and absolute numbers (right) of CD45+ (c) and CD45+CD8+CD90.1+ (d) cells. Expression of PD-1 and LAG-3 (e), Tim-3 and LAG-3 (f), PD-1 and Tim-3 (g) and PD-1, Tim-3 and LAG-3 (h) by CD45+CD8+CD90.1+ pmel-1 cells. The levels of PD-1 expression on pmel-1 cells and their mean fluorescent intensities were compared (i). Bar graphs depict frequencies of PD-1+ (j), LAG-3+ (k), Tim-3+ (l), PD-1+LAG-3+ (m), Tim-3+LAG-3+ (n), PD-1+Tim-3+ (o) and PD-1+Tim-3+LAG-3+ (p) cells in CD45+CD8+CD90.1+ pmel-1 cells. IFNγ (q) and TNFα (r) production by CD45+CD8+CD90.1+ pmel- 1 cells stimulated with or without 1 µg/ml hgp100 peptide assessed by flow cytometry. (s) Ki67 expression in CD45+CD8+CD90.1+ cells. Frequencies (t, v, x) and absolute cell numbers (u, w, y) of IFNγ+(t, u), TNFα+ (v, x), Ki67+ (x, y) cells depicted as bar graphs. Data are representative of 3 experiments with 6 mice per group.
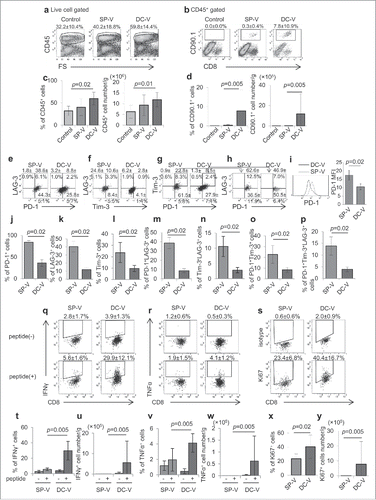
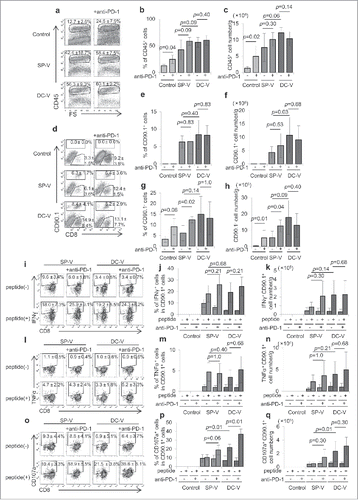
Figure 7. Anti-tumor activity of combining PD-1 signaling blockade with cancer vaccines. C57BL/6 mice (9 mice per group) were treated as described in the legend to . Mice were first inoculated with 1 × 106 B16F10 cells subcutaneously on day 0. On day 5, mice received 1 × 107 naïve pmel-1 transgenic spleen cells. Then mice received no vaccination (a), SP-V (b) or DC-V (c) on days 5 and 12 to these animals and tumor growth monitored. Where indicated, anti-PD-1 antibody (200 µg) was intraperitoneally injected on days 5, 8 and 12. Tumor growth was evaluated every 2–3 days. The graphs show tumor volume of individual mice. The tumor volumes were compared on day 15 (a, b) or 17 (c). Data are representative of two experiments.
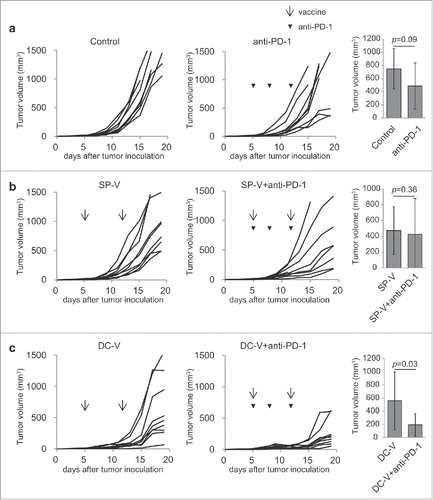
Figure 8. Frequency and functions of vaccine-primed pmel-1 cells with PD-1 signaling blockade. Mice were treated as described in the legend to . On day 19, tumor-infiltrating leukocytes were isolated and examined by flow cytometry; CD45+ cells (a) were further divided into CD45+CD8+CD90.1+ and CD45+CD8+CD90.1− (d) cells. IFNγ (i) and TNFα (l) production and CD107a translocation (o) in CD45+CD8+CD90.1+ pmel-1 cells were assessed by intracellular cytokine staining and CD107a externalization assay, respectively. One plot from each group is depicted. Frequencies (b, e, g, j, m, p) and absolute cell numbers (c, f, h, k, n, q) of CD45+ (b, c), CD45+CD8+CD90.1+ pmel-1 (e, f), CD45+CD8+CD90.1− (g, h), CD45+CD8+CD90.1+IFNγ+ (j, k), CD45+CD8+CD90.1+TNFα+ (m, n) and CD45+CD8+CD90.1+CD107a+ (p, q) cells are depicted by bar graphs. Data are representative of two experiments with 5 mice per group.