Figures & data
Figure 1. Landscape of the CAF composition in LCBM.
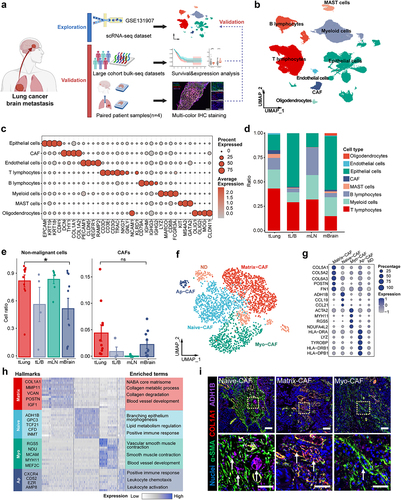
Figure 2. mIF images of paired lung cancer primary/brain metastasis samples.
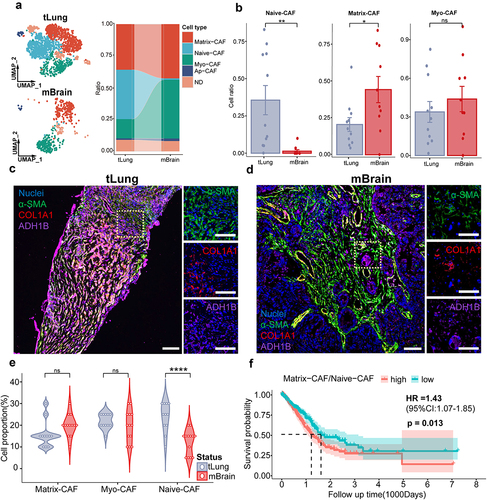
Figure 3. The lineage transition of CAFs in LCBM is associated with hypoxia.
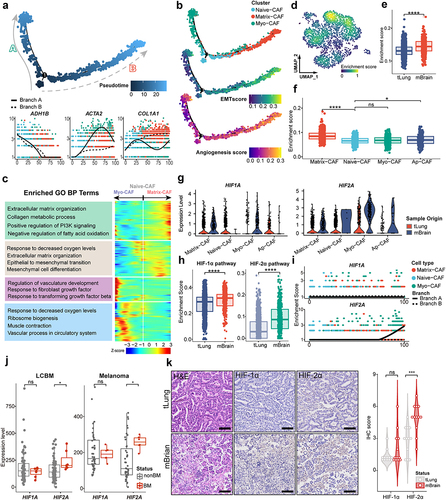
Figure 4. Hypoxia-induced HIF-2α activation drives lineage transition of CAFs.
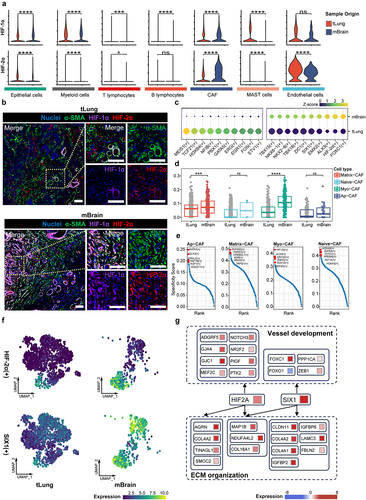
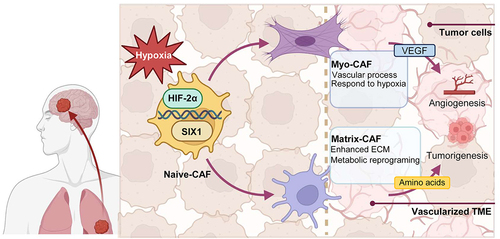
Figure 5. Transited CAFs in LCBM interact with TME and malignant cells.
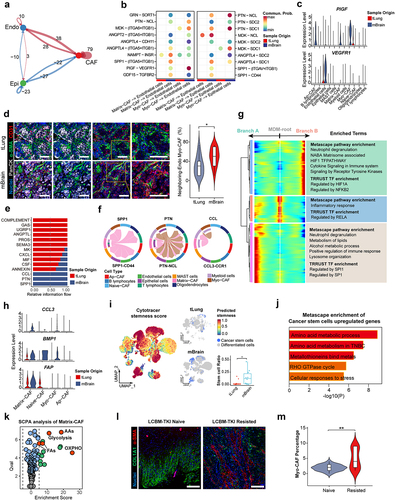
Supplemental Material
Download Zip (2.2 MB)Data availability statement
Raw data of all datasets analyzed in this paper are publicly accessible and can be assessed according to the method given in the paper.