Figures & data
Figure 1. Correlation of increased patients’ overall survival with reduced hnRNP C expression in different cancer types including melanoma.
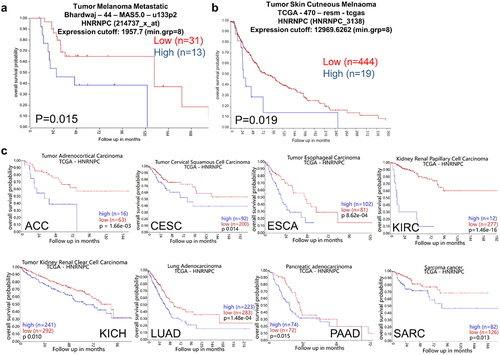
Figure 2. Higher hnRNP C expression levels in tumor samples compared to paired tumor adjacent samples in pan-cancer.
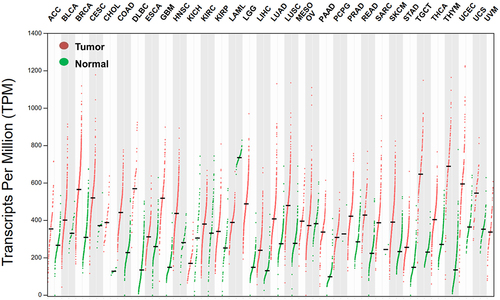
Figure 3. Inverse correlation of hnRNP C and tpn expression in different cancer types.
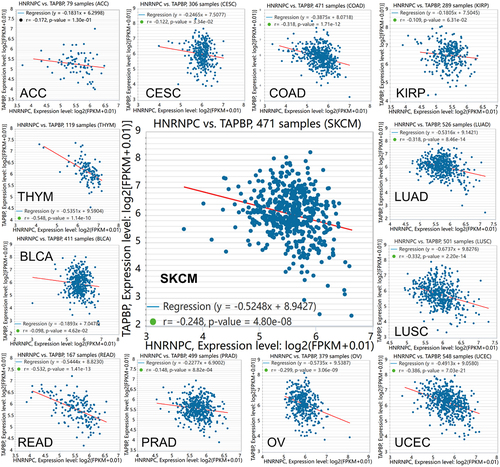
Figure 4. Upregulation of tpn mRNA levels by knockdown of hnRNP C.
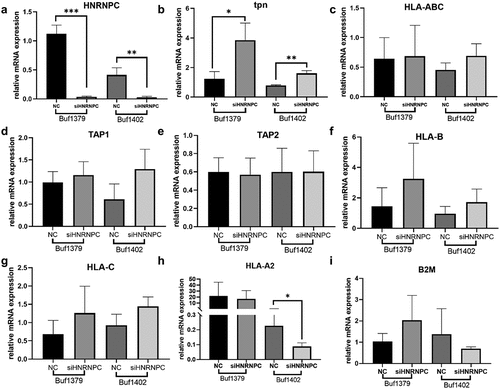
Figure 5. Upregulation of tpn protein levels by knockdown of hnRNP C.
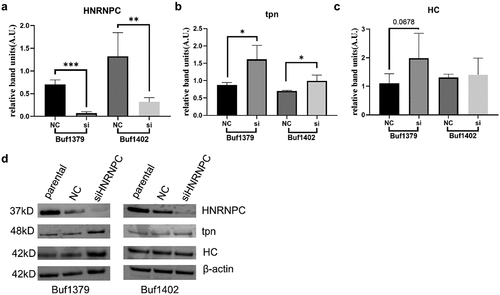
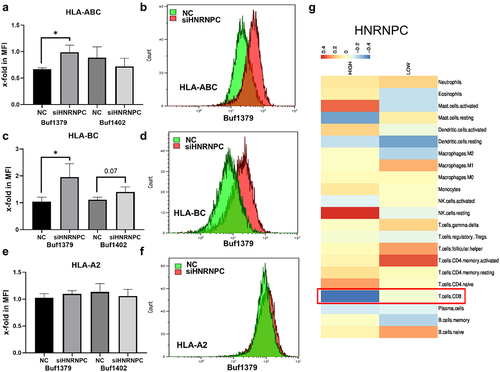
Figure 6. Effect of siRNA-mediated downregulation of hnRNP C on HLA-I cell surface expression and immune cell infiltration.
Supplemental Material
Download MS Word (2.2 MB)Data availability statement
All the data are available in the manuscript or the supplementary materials.
