Figures & data
Table 1. Characteristics of three patients with single-cell analysis.
Figure 1. Phenotypic and TCR clonal analyses of CD8+ T cells from MPE.
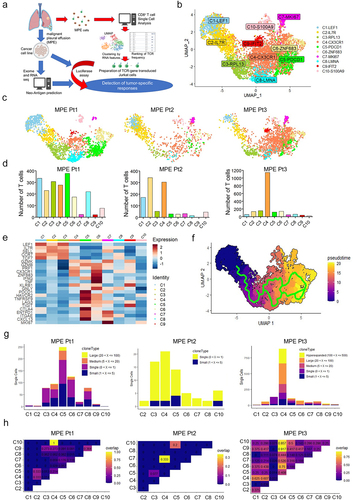
Figure 2. Identification of tumor antigens recognized by CD8+ T cells.
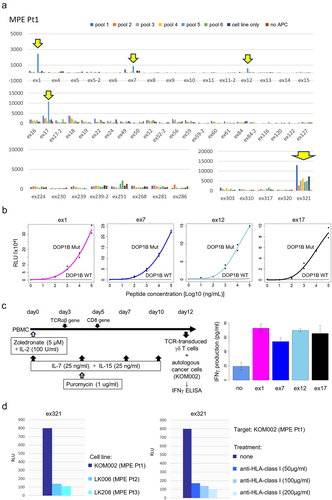
Figure 3. Gene expression analysis of tumor-specific CD8+ T cells from MPE.
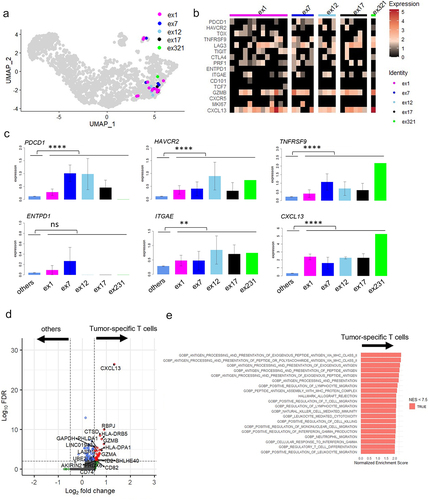
Figure 4. Comparison of tumor specific CD8+ T cells between MPE and TILs.
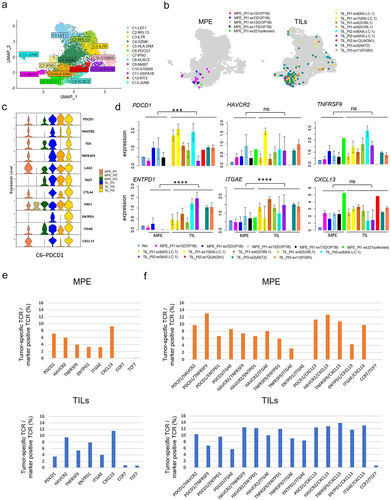
Figure 5. Associations of tumor specific CD8+ T cell fractions in MPE with patients’ prognosis.
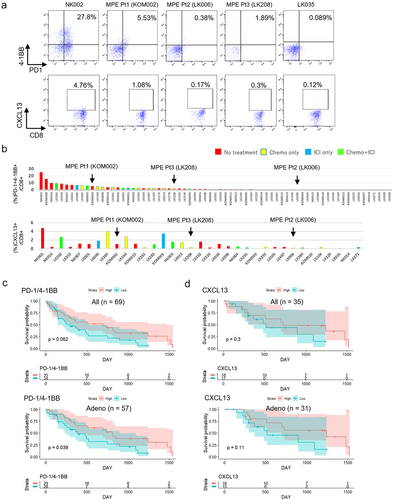
Table 2. Characteristics of patients with flow cytometric analysis.
Supplementary figure.pdf
Download PDF (1.7 MB)Supplementary table.xlsx
Download MS Excel (74.9 KB)Data availability statement
Data are available upon reasonable request.
