Figures & data
Figure 1. Double-negative T (DNT) cells in colorectal cancer (CRC) tissues.
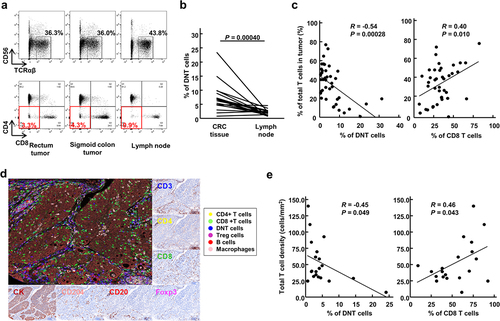
Figure 2. Comparison of TCRβ repertoire among CD4+, CD8+ and double-negative T (DNT) cells in colorectal cancer (CRC) tissues.
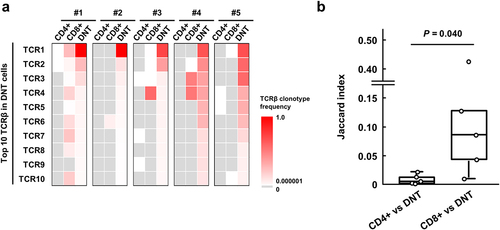
Figure 3. Single-cell transcriptome analysis of CD8+ T and double-negative T (DNT) cells in colorectal cancer (CRC) tissues.
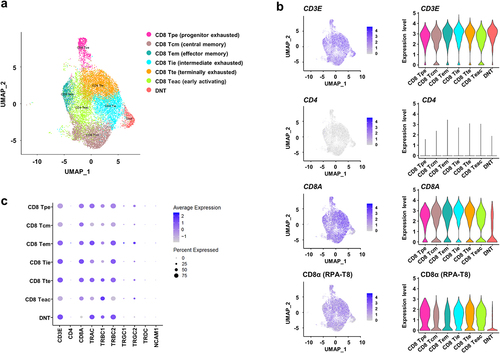
Figure 4. Transcriptional signature of double-negative T (DNT) cells.
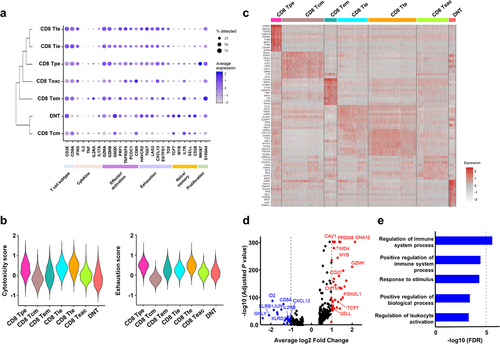
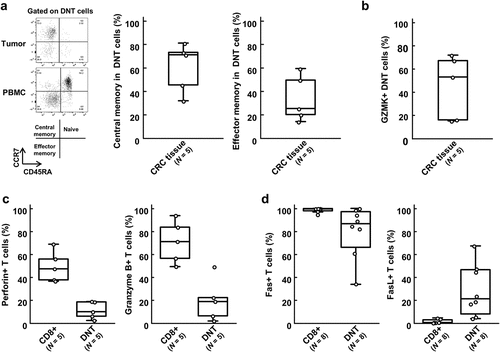
Figure 5. Flow cytometry of specific molecules and cytotoxic molecules in double-negative T (DNT) cells.
Okamura_DNT_suppleTable1_20240527.pdf
Download PDF (176.1 KB)Okamura DNT_suppleFig.docx
Download MS Word (411.1 KB)Data availability statement
All data generated in this manuscript will be provided upon reasonable request to corresponding authors.
