Figures & data
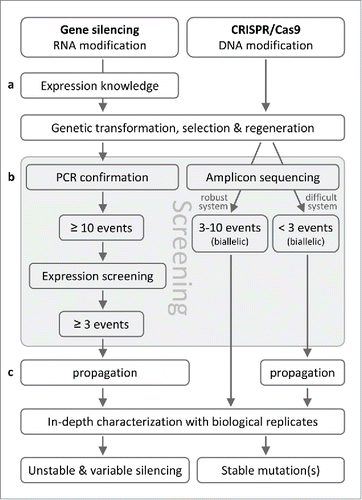
Figure 1. Reddish-brown wood discoloration of 4CL1-edited Populus lines. The photo was modified from Zhou et al. Citation(2015) with permission.
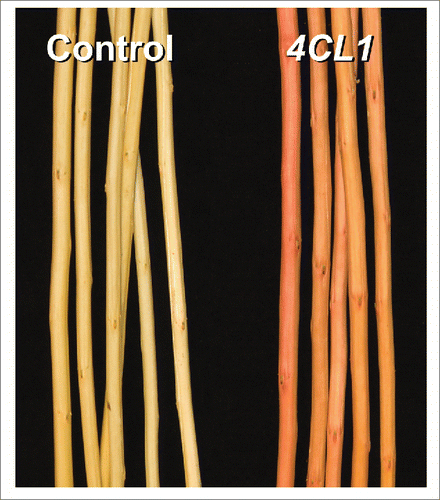
Figure 2. Schematic comparisons of experimental approaches involving gene silencing vs. CRISPR/Cas9 genome editing. (a) For gene silencing by antisense, sense or RNAi approaches, knowledge about the spatiotemporal expression pattern of the target gene is used to select an appropriate promoter for construct preparation. Expression knowledge is not necessary for CRISPR/Cas9 editing at the DNA level, and vectors with a Pol III promoter (e.g., U6 or U3) for gRNA expression and a Cas9 under control of a constitutive (e.g., 35S) promoter are widely applicable. (b) Following regeneration of putative transgenic plants, conventional screening involves PCR confirmation of transgenes followed by expression analysis to select events with desired (or maximum) levels of gene suppression. It is common to screen a large number of events (≥10) to identify a minimum of 3 for subsequent analysis. This process can be greatly simplified with CRISPR/Cas9 mutants, as DNA level analysis by amplicon sequencing can identify events with biallelic mutations for further analysis. No RNA level analysis is necessary. (c) For species with a robust transformation system, multiple biallelic CRISPR/Cas9 events can be used directly as biological replicates for in-depth characterization. In case of difficult-to-transform species for which few biallelic events may be obtained, micropropagation is necessary to obtain biological replicates before in-depth characterization, as is typically done using gene silencing.