Figures & data
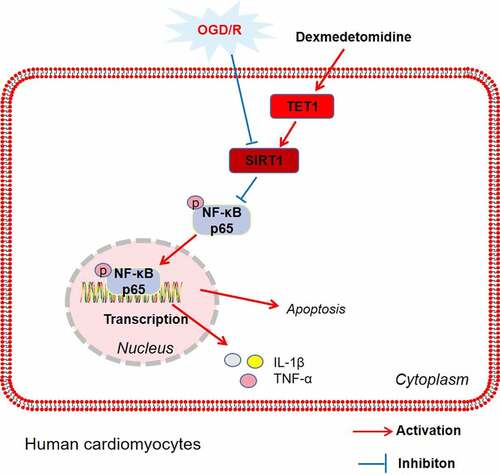
Figure 1. DEX inhibited OGD/R – mediated myocardial injury.
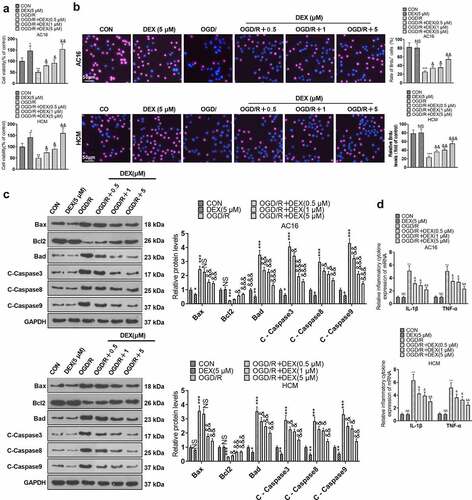
Figure 2. DEX facilitated Sirt1 and inactivated NF-κB.
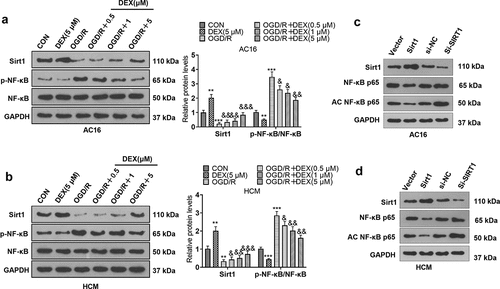
Figure 3. Inhibiting Sirt1 attenuated DEX-mediated myocardial protection.
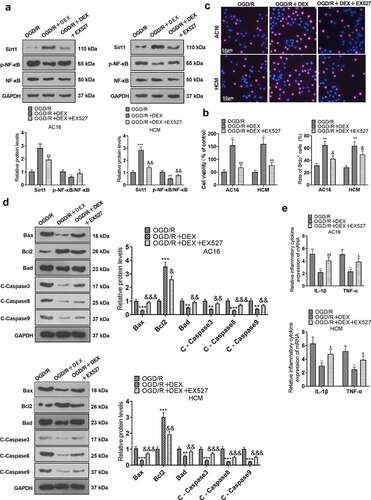
Figure 4. DEX boosted TET1 and abated OGD/R-mediated DNA methylation in cardiomyocytes.
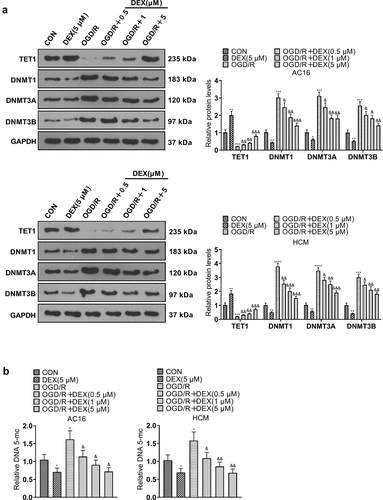
Figure 5. TET1 knockdown impeded the DEX-mediated myocardial protective effect.
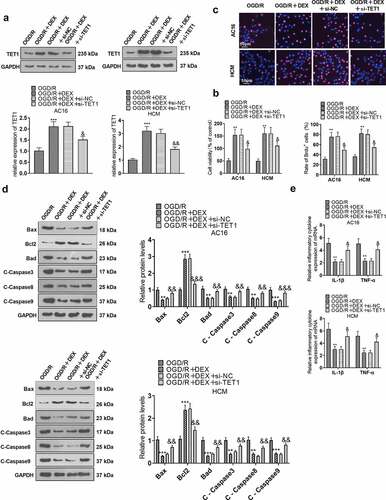
Figure 6. TET1 mediated demethylation of the Sirt1 promoter and lifted Sirt1 expression.
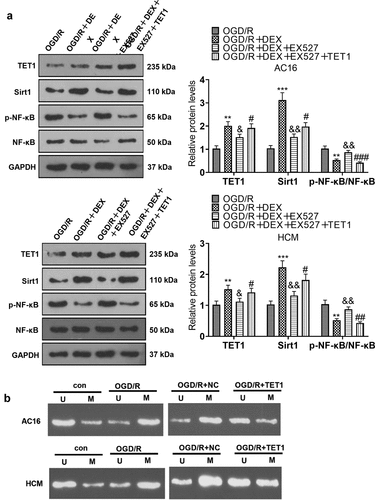
Figure 7. Knockdown of TET1 weakened DEX-mediated myocardial protection.
Data availability statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
