Figures & data
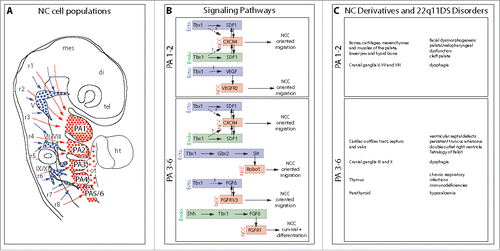
Figure 1. The SDF1/CXCR4 signaling pathway: A new player involved in DiGeorge/22q11-deletion syndrome malformations. A. Schematic representation of the different NC cell populations that colonize the pharyngeal arches, the cardiac outflow tract and the cranial ganglia. NC cells that express the CXCR4 receptor and respond to SDF1 signals allowing them to migrate along the superficial ectoderm and populate the arches and the cardiac outflow tract are indicated in red. Those which do not express CXCR4 and migrate ventrally to contribute to the cranial sensory ganglia are in blue. Arrows indicate the level of origin of NC cells along the rostro-caudal axis. tel, telencephalon; di, diencephalon; mes, mesencephalon; r1-r8, rhombomeres 1-8; V, trigeminal ganglion; VII/VIII, facial and vestibuloacoustic ganglia; IX/X, glossopharyngeal and vagal ganglia; PA1-6, pharyngeal arches 1-6; ht, heart; ot, outflow tract. B. Putative signaling pathways regulating migration, survival and differentiation of NC cells in PA1-2 and PA3-6, respectively. The tissular origin of the signal (enodermal or ectodermal) is indicated. Single arrows with plain line represent demonstrated (direct or indirect) regulations and double arrows indicate reciprocal regulations. Arrows with dashed line and associated with question marks represent hypothetical regulatory pathways. C. Lists of the main NC derivatives generated in PA1-2 and PA3-6, respectively, and of the corresponding disorders commonly observed in 22q11DS. Neural defects consecutive to CXCR4 signaling defects and causing mental retardation and other psychiatric disorders are not listed because, to our knowledge, they are not directly related with anomalies in pharyngeal NC cells.