Figures & data
Figure 1. Schematic representation of the energy landscape of an ordered protein (top panel) and a close-up view of the bottom of the funnel-like energy landscape of a protein containing ordered structural elements (shown in white) and flexible regions (shown in red). Modified with the permission from Burger, V.M.; Gurry, T.; Stultz, C.M. Intrinsically Disordered Proteins: Where Computation Meets Experiment. Polymers 2014, 6, 2684–2719 (ref.Citation45), which is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. (CC BY 4.0).
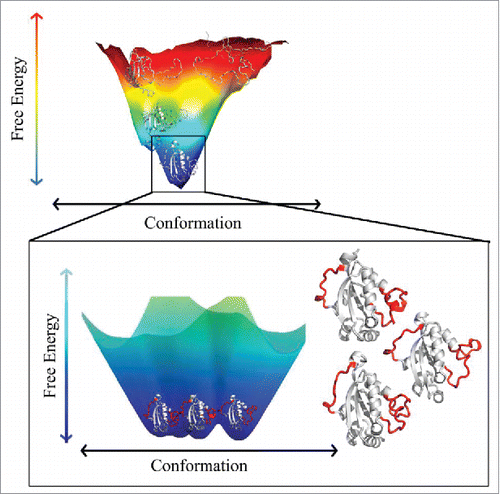
Figure 2. Characterization of structural flexibility of 6 proteins considered as crystallization classics. For each protein, structural alignments were conducted using the MultProt (http://bioinfo3d.cs.tau.ac.il/MultiProt/)Citation64 for pairs of structures characterized by the largest difference according to the PDBFlex analysis. Analyzed structures include PDB entries 105mA (blue ribbon) and 2eb8A (green ribbon)Citation65 for myoglobin, 1lkrB (blue ribbon) and 1a2yC (green ribbon) for lysozyme, 1f0vB (blue ribbon)Citation62 and 3fkzA (green ribbon)Citation66 for RNase A, 1oxgA (blue ribbon)Citation67 and 1ex3A (green ribbon)Citation68 for chymotrypsin, 1u75B (blue ribbon)Citation69 and 3nbsC (green ribbon)Citation63 for cytochrome c, and 2abzB (blue ribbon)Citation70 and 1hdqA (green ribbon)Citation71 for carboxypeptidase A1. Structures presented in this plot were generated using a molecular graphics program VMD.Citation72
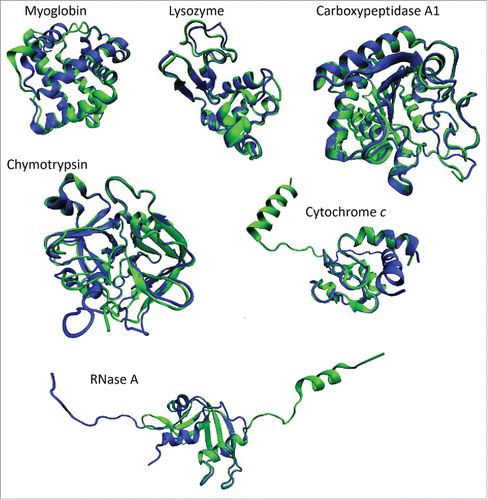
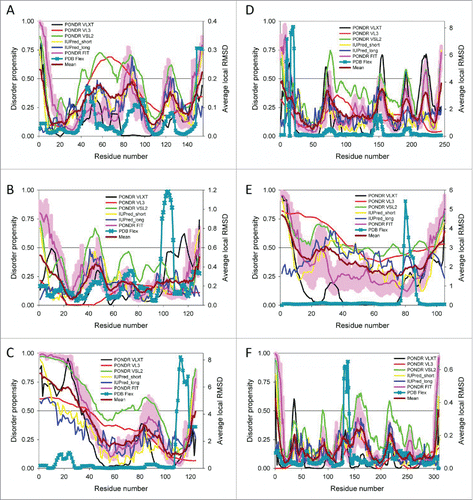
Figure 3. Comparison of the PDBFlex-evaluated structural flexibility and intrinsic disorder propensity of 6 crystallization classics: A. Sperm whale myoglobin (UniProt ID: P02185); B. Hen egg white lysozyme (UniProt ID: P00698); C. Bovine pancreatic ribonuclease A (UniProt ID: P61823); D. Bovine chymotripsin (UniProt ID: P00766); E. Horse cytochrome c (UniProt ID: P00004); and F. Bovine carboxypeptidase A1 (UniProt ID: P00730). For each protein, the PDBFlex-evaluated structural flexibility is shown in a form of sequence distribution of local average RMSD values (dark cyan curves with crosses). Disorder profiles were generated by the superposition of the outputs of PONDR® VLXT, PONDR® VL3, PONDR® VSL2, and PONDR® FIT,Citation73-78 as well as the IUPred web server,Citation79 IUPred_short and IUPred_long. The corresponding outputs are shown by black, red, green, pink, yellow, and blue lines, respectively. In each plot, dark red line shows the mean disorder propensity calculated by averaging disorder profiles of individual predictors. The light pink shadow around the PONDR® FIT shows error distribution. In these analyses, the predicted intrinsic disorder scores above 0.5 are considered to correspond to the disordered residues/regions, whereas regions with the disorder scores between 0.2 and 0.5 are considered flexible.