Figures & data
Table 1. Primer sequences employed in this project.
Figure 1. Morphologic changes in the cells on TCPS at different time periods of differentiation. (a) Day 0, the cells were spindle shaped before induction (original magnification 100×); (b) Day 1, the induced cells turned shorter and changed into round cells and islet-like cell (ILC) clusters appeared (original magnification 100×); and (c) Day 6, the center of large cell cluster was dense and its peripheral migrated to outer part of cluster; (d) large ILC separated to small cell clusters. Scale bar 100 uM.
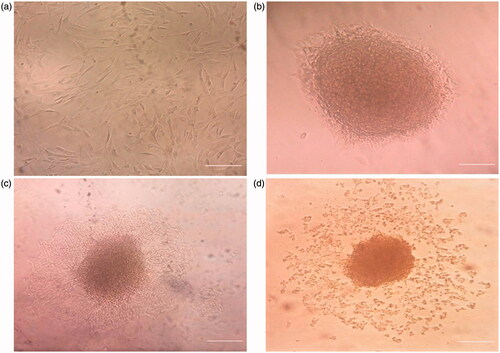
Figure 3. Immunostaining of CJMSCs-derived IPCs. The cells were seeded on 3D scaffold and TCPS were then maintained in induction medium for 16 days and were analyzed for expression of insulin, c-peptide, glucagon and Pdx-1 markers. Cells were co-stained with 4,6-diamidino-2-phenylindole to visualize nuclei (blue). Scale bar 100 μM).
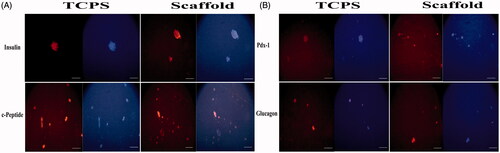
Figure 4. Flow cytometry analysis of insulin, c-peptide, glucagon and Pdx-1 proteins in CJMSCs-derived IPCs on TCPS.
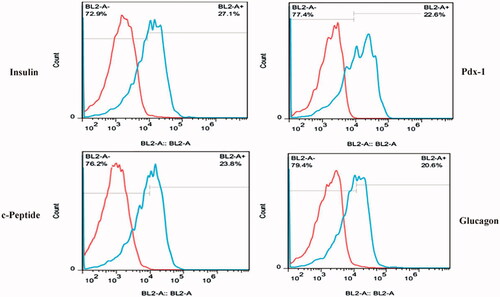
Figure 5. In vitro functional (insulin ELISA) assay in response to different concentrations of glucose.
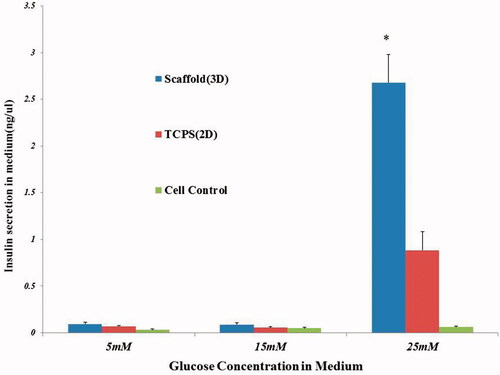
Figure 6. Gene expression profile of cells differentiated on P-PCL and TCPS on day 7 and 16. The cells were maintained in induction medium for 7 and 16 days and analyzed for expression of pancreatic markers. The column ratio is the expression rate of genes compared with untreated cells. GAPDH is shown as a control for RNA sample quality. Rest software was used for gene expression analysis using real-time PCR data. Asterisks show significance expression rate. *p ≤ .05.
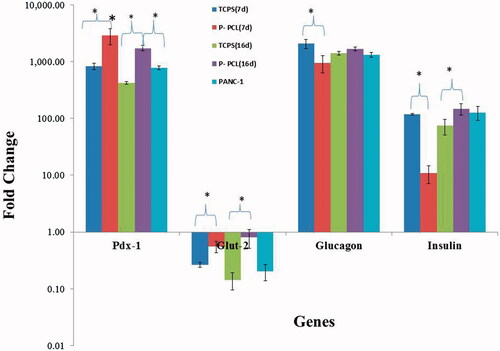
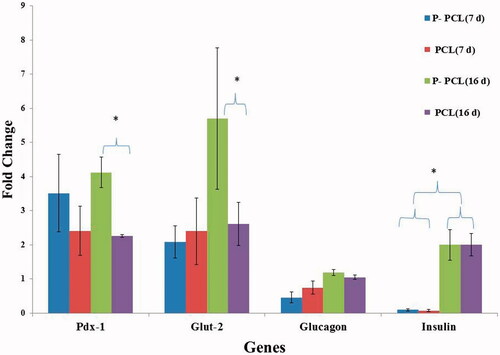
Figure 7. Gene expression profile of cells differentiated on plasma treated PCL and untreated PCL scaffold on day 7 and 16. The cells were maintained in induction medium for 7 and 16 days and analyzed for expression of pancreatic markers. The column ratio is the expression rate of genes compared with cells differentiated on TCPS. GAPDH are shown as a control for RNA sample quality. Rest software was used for gene expression analysis using real-time PCR data. Asterisks show significance expression rate. *p ≤ .05.