Figures & data
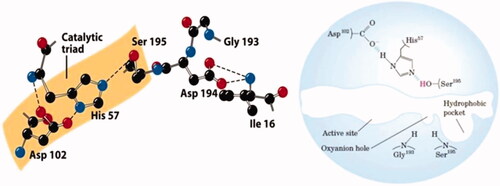
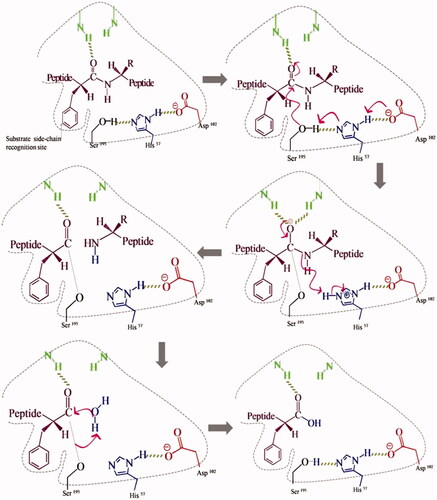
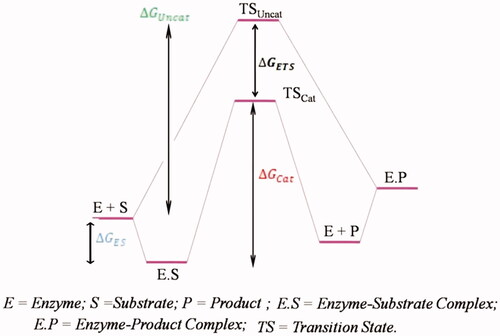
Table 1. Summary of analytical techniques for rational MIP assessment.
Table 2. Molecularly imprinted catalysts versus enzymes.
![Figure 8. Esterase MIP with the imprints of substrate analogue as chymotrypsin mimic [Citation46].](/cms/asset/9b673a9c-cd6e-40c6-b4a6-fe3ce9807012/ianb_a_1576703_f0008_c.jpg)
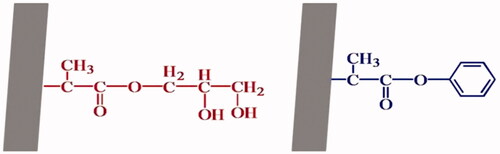
![Figure 10. Hydrolysis of p-nitrophenyl acetate [Citation47].](/cms/asset/79c5a976-d86b-4c27-bd1f-29a61405f73c/ianb_a_1576703_f0010_c.jpg)
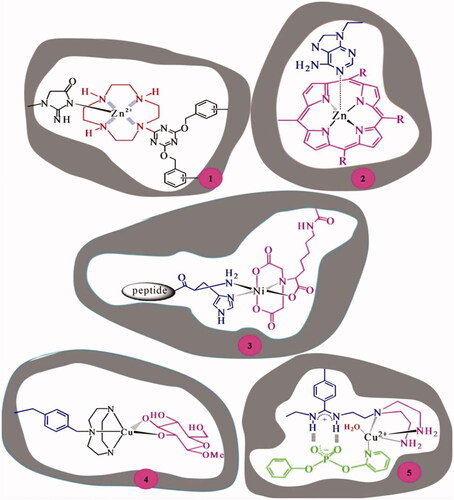
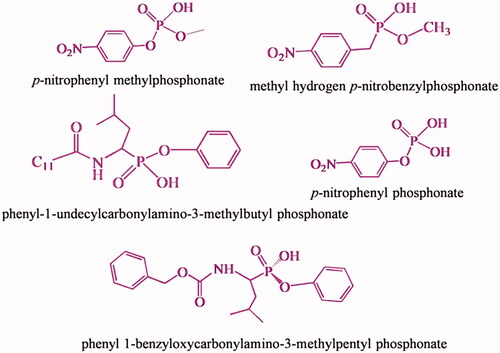
![Figure 11. Homogeneous and heterogeneous esterase MIPs with the imprints of methyl hydrogen p-nitrobenzylphosphonate [Citation52].](/cms/asset/98e6db65-1ee8-4f1a-a6ab-444fc754a7e2/ianb_a_1576703_f0011_c.jpg)
![Figure 12. Heterogeneous esterase MIP [Citation52].](/cms/asset/e2f30035-0418-4198-a59b-c3c54f39af59/ianb_a_1576703_f0012_c.jpg)
![Figure 13. Water soluble homogeneous esterase MIP with the imprints of phenyl-1-enzyloxycarbonylamino-3-methylpentylphosphonate [Citation48].](/cms/asset/48654b84-aa81-4fb1-81f2-60b6500fdd75/ianb_a_1576703_f0013_c.jpg)
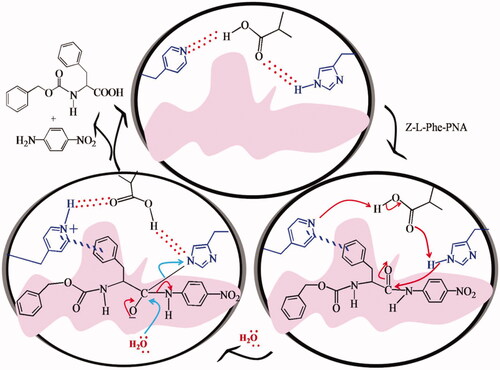
![Figure 15. Esterase MIP with the imprints of chiral phosphonate analogue of phenylalanine as TSA [Citation6].](/cms/asset/6f5f3e5f-0ec8-4444-ba22-5d303e59ec06/ianb_a_1576703_f0015_c.jpg)
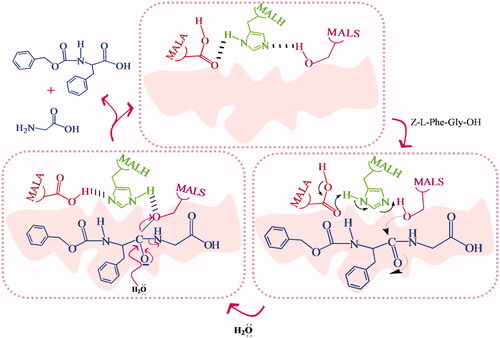
![Figure 16. Esterase MIP synthesized using phenyl 1-benzyloxycarbonylamino-3-methylpentylphosphonate TSA [Citation54].](/cms/asset/e4c63635-4ae4-41e1-8b84-481963c1d2ff/ianb_a_1576703_f0016_c.jpg)
![Figure 17. Esterase MIP synthesized using N- (N-benzyloxycarbonyl-L-leucinoyl) anthranilic acid GSA [Citation54].](/cms/asset/6c583200-ca33-480a-a7bf-d95d96ab5fdb/ianb_a_1576703_f0017_c.jpg)
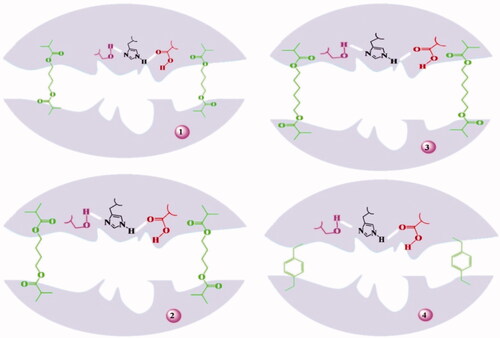
![Figure 18. Water soluble and water insoluble MIPs [Citation55].](/cms/asset/3089db87-dd25-4de6-b6c6-feb10af8d62e/ianb_a_1576703_f0018_c.jpg)
![Figure 19. Water soluble esterase MIP’ with the imprints of phenyl-1-benzyloxycarbonyl-3-methylpentyl phosphonate [Citation56].](/cms/asset/4b1a1be6-304e-4107-8e65-13959f77f38b/ianb_a_1576703_f0019_c.jpg)
![Figure 20. Stoichiometric non-covalent imprinting strategy employing a monomer and bifunctional TSA phosphonate ester [Citation57].](/cms/asset/1eb3af7a-56eb-4418-b448-9ab91516b846/ianb_a_1576703_f0020_c.jpg)
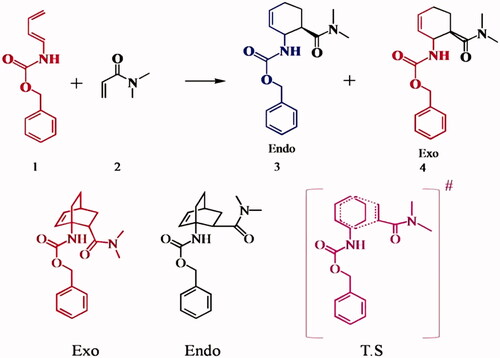
![Figure 21. First reported chymotrypsin mimc [Citation41,Citation42].](/cms/asset/59df08a4-9dbd-410a-9829-a55312400a83/ianb_a_1576703_f0021_c.jpg)
![Figure 23. Imidazole-containing DVB crosslinked esterase MIP for the esterolysis of p-nitrophenyl acetate [Citation51].](/cms/asset/a0cb9e82-9354-4214-89cb-7241a5e7dd60/ianb_a_1576703_f0023_c.jpg)
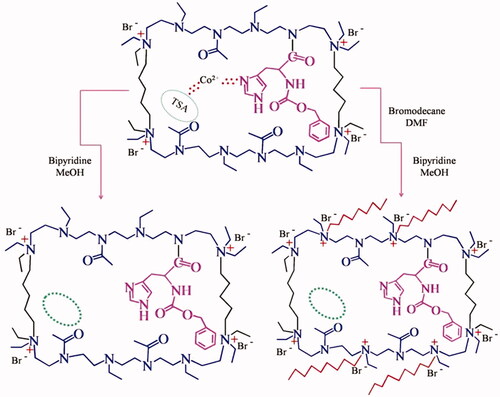
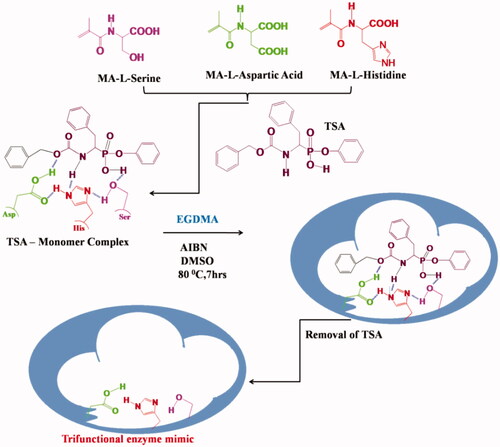
![Figure 24. Polymer catalyst with TSA and the imprints of TSA [Citation59, Citation60].](/cms/asset/7047c5cb-ba07-4c89-9c1a-90aeb5a5604b/ianb_a_1576703_f0024_c.jpg)
![Figure 25. Preparation of a TSA imprinted catalyst by labile covalent binding and non-covalent binding [Citation59,Citation60].](/cms/asset/a92dad48-0f7d-40c4-9d05-189e42fbb855/ianb_a_1576703_f0025_c.jpg)
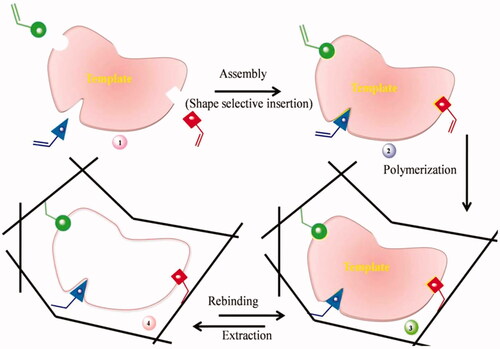
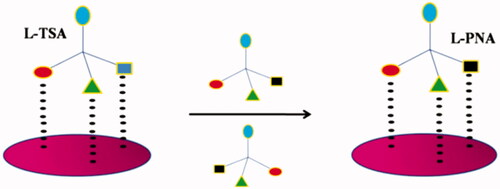
![Figure 26. Molecular imprinting using template monomer [Citation58].](/cms/asset/f277d2d5-d034-4c57-8a8f-bedc66b0cb2f/ianb_a_1576703_f0026_c.jpg)
![Figure 27. Esterolysis of long chain ester [Citation50].](/cms/asset/1a03422f-bdf6-4b5b-969d-ff173eac0f44/ianb_a_1576703_f0027_c.jpg)
![Figure 28. Introduction of nucleophilic 4-(N,N-dimethylamino)pyridines as functional monomer in imprinting [Citation61].](/cms/asset/6f65a525-75e7-4b97-bd5b-e7418ae40330/ianb_a_1576703_f0028_c.jpg)