Figures & data
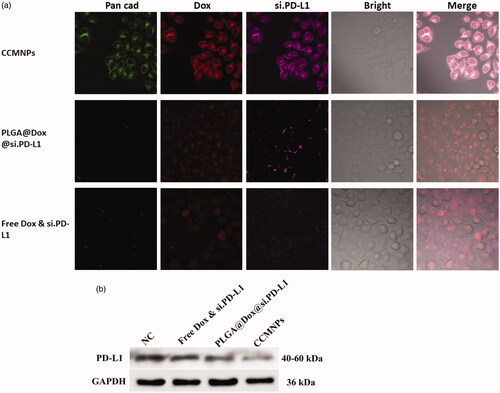
Figure 1. Characterization of CCMNPs. (a) Size intensity curves of bare PLGA cores, c, cancer cell membrane vesicles, and CCMNPs measured by dynamic light scattering (DLS). (b) Hydrodynamic size of bare PLGA cores, PLGA@[email protected], cancer cell membrane vesicles, and CCMNPs. Bars represent means ± SD (n = 3). (c,d) Transmission electron micrographs of (c) PLGA@[email protected] and (d) CCMNPs. Samples were negatively stained with phosphotungstic acid. All scale bars = 100 nm.
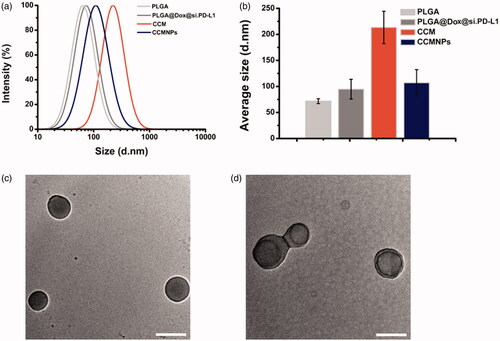
Figure 2. Membrane antigen characterization of CCMNPs. (a) SDS-PAGE protein analysis of cancer cell membrane vesicles and CCMNPs. Samples were run at equal protein concentration and stained with Coomassie Blue. (b) Western blotting analysis for membrane-specific protein, pan-cadherin.
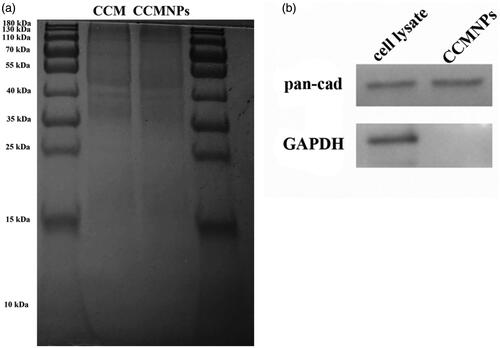
Figure 3. Targeted delivery of CCMNPs based on self-recognition. (a) Flow cytometric analysis of Hela cells incubated with blank solution, free Dox, PLGA@[email protected] or CCMNPs. (b) Flow cytometric analysis of MDA-MB-231 cells incubated with blank solution, free Dox, PLGA@[email protected] or CCMNPs. (c) Cytotoxicity profiles in Hela and MDA-MB-231 cells obtained after 48 h incubation of PLGA, PLGA@[email protected], CCM or CCMNPs. *p < .05, and ***p < .001.
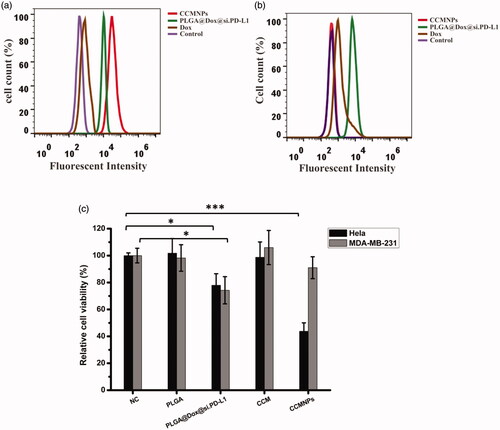
Figure 4. CCMNPs serve as a homotypically targeted delivery vehicle. (a) Confocal fluorescent imaging of Hela cells incubated with CCMNPs, PLGA@[email protected] or free Dox & si.PD-L1. (b) PD-L1 knockdown was induced after CCMNPs incubation for 48 h.