Figures & data
Figure 1. Characterization of targeting liposomes. (A) MALDI-TOF-MS spectrum of DSPE-PEG2000-NHS. (B) MALDI-TOF-MS spectrum of DSPE-PEG2000-R8.
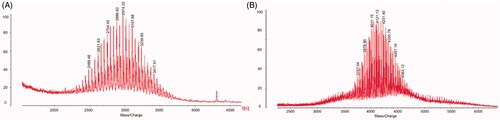
Figure 2. Characterizations of R8 modified epirubicin–dihydroartemisinin liposomes. (A) AFM image of R8 modified epirubicin–dihydroartemisinin liposomes. (B) TEM image of R8 modified epirubicin–dihydroartemisinin liposomes. (C) Particle size of R8 modified epirubicin–dihydroartemisinin liposomes. (D) Zeta potential of R8 modified epirubicin–dihydroartemisinin liposomes.
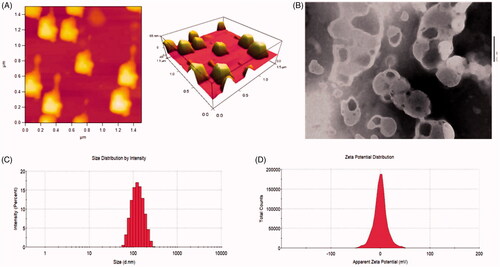
Table 1. Characterization of liposomes.
Figure 3. Intracellular uptake and targeting effects after incubation with varying formulations. (A) Cellular uptake of A549 cells. (B) Fluorescence intensity of epirubicin in A549 cells. (C) Fluorescence microscopy images of A549 cells incubated with varying formulations. (a) Blank control; (b) epirubicin liposomes; (c) epirubicin–dihydroartemisinin liposomes; (d) R8 modified epirubicin–dihydroartemisinin liposomes; p < .05. 1, vs. Blank control; 2, vs. epirubicin liposomes; 3, vs. epirubicin–dihydroartemisinin liposomes.
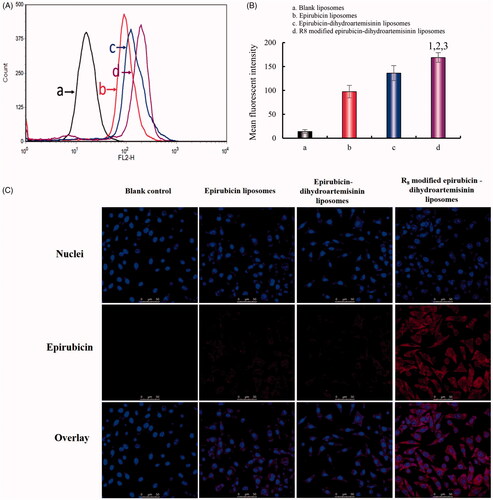
Figure 4. In vitro release rates of drugs from varying liposomes in the simulated body fluids. (A) Release rate of epirubicin from varying formulations in PBS solution containing 10% mouse plasma; (B) release rate of epirubicin from varying formulations in normal saline; (C) release rate of dihydroartemisinin from varying formulations in PBS solution containing 10% mouse plasma; (D) Release rate of dihydroartemisinin from varying formulations in normal saline. Data are presented as mean ± standard deviation (SD) (n = 3).
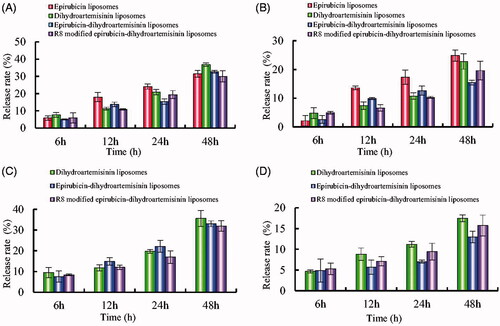
Figure 5. Inhibitory effects on A549 cells after treatments with the varying formulations. (A) Inhibitory effects of free drugs; (B) Inhibitory effects of liposomal formulations. p < .05; I, vs. free dihydroartemisinin; II, vs. free epirubicin; III, vs. dihydroartemisinin: epirubicin = 1:1; IV, vs. dihydroartemisinin : epirubicin = 1:5; a, vs. blank liposomes; b, vs. dihydroartemisinin liposomes; c, vs. epirubicin liposomes; d, vs. epirubicin–dihydroartemisinin liposomes; data are presented as mean ± SD (n = 4).
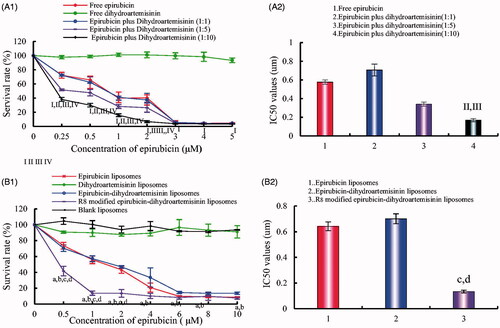
Figure 6. Destructive effects on VM channels and blocking effects on A549 cells migration in vitro after treatment with varying formulations. (A) Blocking effects on A549 cells migration; (B) destructive effects on VM channels, (a) blank control; (b) epirubicin liposomes; (c) dihydroartemisinin liposomes; (d) epirubicin–dihydroartemisinin liposomes; (e) R8 modified epirubicin–dihydroartemisinin liposomes.
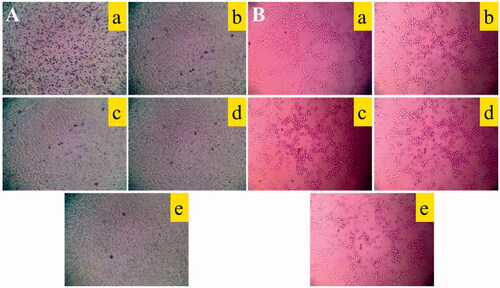
Figure 7. Blocking wound-healing effects in A549 cells after treatments with the varying liposomal formulations.
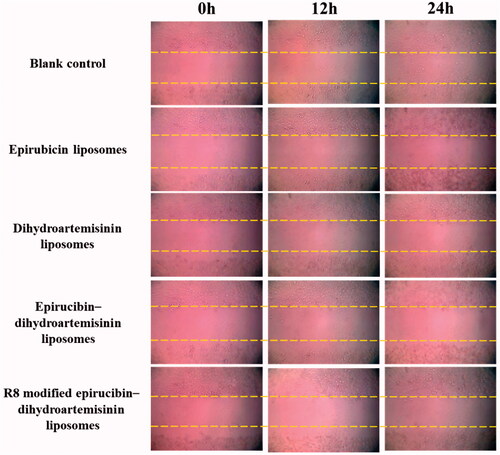
Figure 8. Adhesion rate of A549 cells on matrigel after incubation with the varying formulations. Adhesion rate of A549 cells on matrigel. p < .05; (1) blank liposomes; (2) epirubicin liposomes; (3) dihydroartemisinin liposomes; (4) epirubicin–dihydroartemisinin liposomes; (5) R8 modified epirubicin–dihydroartemisinin liposomes. a, vs. Blank liposomes.
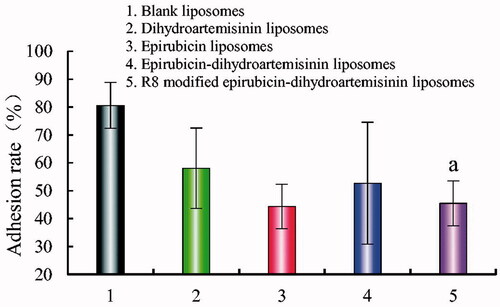
Figure 9. In vivo real-time imaging observation after intravenous administration and antitumor effects in A549 cells xenografts mice after treatments with the varying formulations. (A) In vivo real-time imaging observation after intravenous administration of the varying formulations, (B) body weight changes, (C) tumor volume changes.
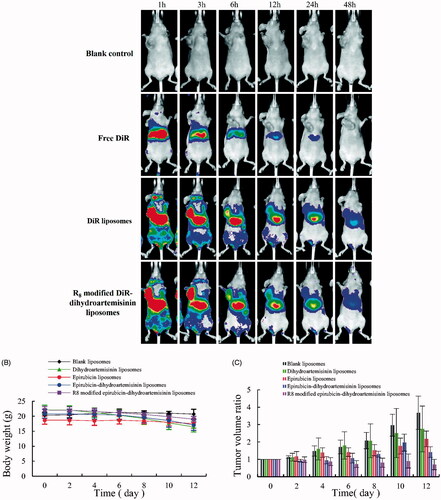
Figure 10. Regulating effects on VM related proteins. (A) MMP-2 protein expression ratio, (B) VE-Cad protein expression ratio, (C) TGF-β1 protein expression ratio, (D) HIF-1α protein expression ratio. Data are presented as the mean ± SD (n = 3). (1) Blank liposomes; (2) epirubicin liposomes; (3) dihydroartemisinin liposomes; (4) epirubicin–dihydroartemisinin liposomes; (5) R8 modified epirubicin–dihydroartemisinin liposomes. p<.05. a, vs. Blank liposomes; c, vs. Dihydroartemisinin liposomes. MMP-2: matrix metalloproteinase-2; VE-Cad: VE-Cadherin; TGF-β1: transforming growth factor-β1; HIF-1α: hypoxia inducible factor-1α.
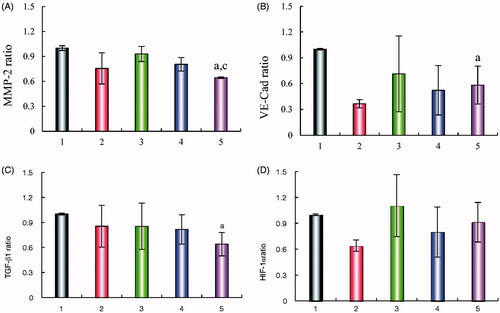
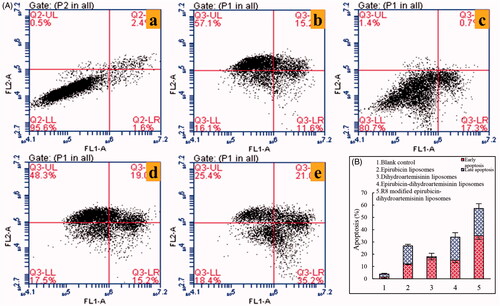
Figure 11. Apoptosis inducing effects on A549 cells after incubation with the varying formulations. (A) Plots by flow cytometry of apoptosis-inducing effects in A549 cells; (B) apoptosis-inducting effects after treatments with the varying formulations. Data are presented as the mean ± SD (n = 3). (1) Blank liposomes; (2) epirubicin liposomes; (3) dihydroartemisinin liposomes; (4) epirubicin–dihydroartemisinin liposomes; (5) R8 modified epirubicin–dihydroartemisinin liposomes.