Figures & data
Figure 1. JHDM1A was acetylated by p300 in osteosarcoma cells. (A) MG-63 and HOS cells were transfected with plasmids expressing Flag-tagged JHDM1A, respectively. Immunoprecipitation (IP) assay with anti-Flag antibody and immunoblotting assay with pan-acetyl-lysine (Ac-K) antibody showed that JHDM1A could be acetylated in osteosarcoma MG-63 and HOS cells. TSA and NAM further promoted the acetylation of JHDM1A. (B) p300 and Flag-JHDM1A were co-transfected into HEK293 cells. IP assay with anti-Flag antibody and immunoblotting assay with Ac-K antibody showed that p300 induced JHDM1A acetylation. (C) sh-p300 was transfected into MG-63 and HOS cells. IP assay with anti-Flag antibody and immunoblotting assay with Ac-K antibody showed that knockdown of p300 reduced the acetylation of JHDM1A. (D) IP assay was carried out in HOS cell lysates using antibody against p300. Western blotting using JHDM1A antibody showed that JHDM1A also existed in the complex. (E) HA-tagged JHDM1A and Myc-tagged p300 were co-transfected into MG-63 cells. IP assay was carried out in MG-63 cell lysates using antibody against HA. Western blotting using anti-Myc antibody and anti-HA showed that p300 also existed in the complex. (F) Glutathione S-transferase (GST) pull down assay with GST-tagged JHDM1A and p300 showed that p300 directly interacted with JHDM1A.
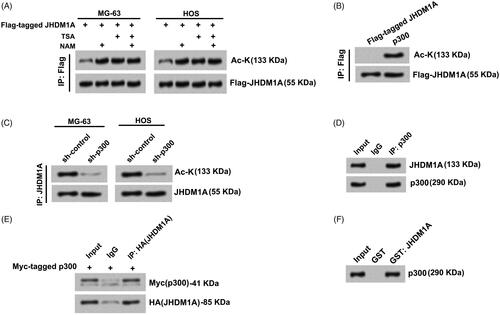
Figure 2. p300 directly acetylated JHDM1A at lysine residues 409 (K409). (A) K409 site of JHDM1A was replaced using arginine (R) to form acetylation-resistant (K-R mutated) JHDM1A. p300 immuno-precipitates (from Flag-p300 expressed in HEK293 cells) and recombinant full-length wild-type (WT) JHDM1A protein or K-R mutated JHDM1A protein (from bacteria) were co-incubated. Immunoblotting assay with pan-acetyl-lysine (Ac-K) antibody showed that the K-R mutated JHDM1A was resistant to p300-mediated acetylation. (B) K409 site of JHDM1A was replaced using glutamine (Q) to form acetylation-mimicking (K-Q mutated) JHDM1A. p300 and Flag-tagged WT (or K-R, K-Q mutated) JHDM1A were co-transfected into HEK293 cells. Immunoprecipitation (IP) assay with anti-Flag antibody and immunoblotting assay with Ac-K antibody showed that both K-R JHDM1A and K-Q JHDM1A were resistant to p300-mediated acetylation.
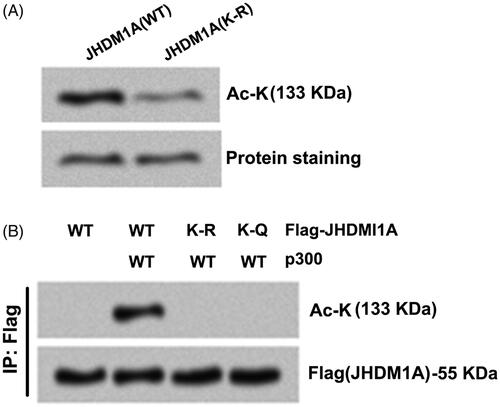
Figure 3. Acetylation of JHDM1A disrupted its binding to nucleosomes and demethylation of H3K36me2. (A) Recombinant core histones and biotin-labeled DNA (Bio-nucl) were utilized for reconstituting nucleosome. WT or K-Q mutated JHDM1A was mixed with reconstituted nucleosomes. Immunoblotting assay with anti-JHDM1A antibody showed that the binding of K-Q mutated JHDM1A to nucleosomes was reduced in vitro. (B) WT or K-Q, K-R mutated JHDM1A was incubated with bulk histone. Immunoblotting assay with anti-H3K36me2 antibody, anti-H3K36me3 antibody or anti-JHDM1A antibody showed that the acetylation status of JHDM1A had no influences on its ability to demethylate H3K36 in bulk histones in vitro. H3K36me3 level was used as a loading control. (C) WT or K-Q, K-R mutated JHDM1A was incubated with mononucleosomes isolated from MG-63 cells. Immunoblotting assay with anti-H3K36me2 antibody, anti-H3K36me3 antibody or anti-JHDM1A antibody showed that the K-Q mutated JHDM1A could not demethylate H3K36 in mononucleosomes. H3K36me3 level was used as a loading control.
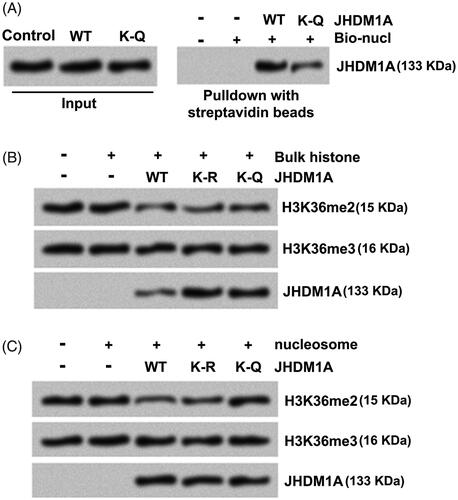
Figure 4. Acetylation of JHDM1A impaired its ability to repress p21 and puma transcription. (A and B) Bindings of Flag-tagged WT or K-R, K-Q mutated JHDM1A proteins to the p21 and puma promoters were evaluated by ChIP analysis with anti-Flag antibodies. The bindings between K-Q mutated JHDM1A and p21 or puma promoters were notably reduced. (C and D) Followed by Flag-tagged WT or K-R, K-Q mutated JHDM1A transfection, the bindings of H3K36m2 to the p21 and puma promoters were measured by ChIP analysis. K-Q mutated JHDM1A failed to reduce the binding H3K36m2 to the p21 and puma promoters (E and F) Followed by Flag-tagged WT or K-R, K-Q mutated JHDM1A transfection, the p21 and puma mRNA levels in MG-63 cells were tested by real-time RT-PCR. (G) The possible schematic presentation of chip regions and PCR primer binding sites. *p ˂ .05; **p ˂ .01.
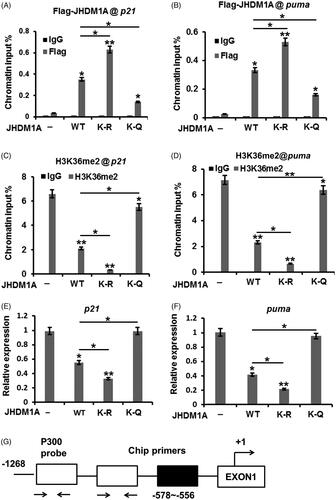
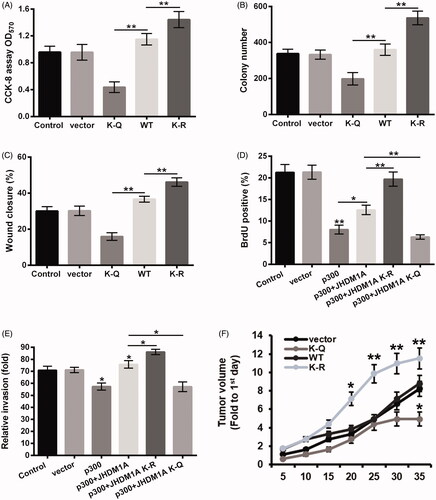
Figure 5. Acetylation of JHDM1A inhibited osteosarcoma carcinogenesis. Plasmid expressed WT, K-Q or K-R mutated JHDM1A was transfected into HOS cells, respectively. Empty plasmid was used as a negative control. (A) Cell viability, (B) colony formation and (C) cell invasion were tested using CCK-8 assay, colony formation assay and wound closure assay, respectively. Plasmid expressed p300 and plasmid expressed WT, K-Q or K-R mutated JHDM1A were co-transfected into HOS cells, respectively. Empty plasmid was used as a negative control. (D) Cell proliferation and (E) cell invasion were tested using 5-bromo-2′-deoxyuridine (BrdU) incorporation assay and two chamber transwell assay, respectively. (F) HOS cells were transfected with HA-tagged WT, K-Q or K-R mutated JHDM1A-expressing plasmids and injected into the hind limbs of mice to form mouse tumor models. The tumor volumes were measured every 5 days after injection. *p ˂ .05; **p ˂ .01.