Figures & data
Table 1. The clinical feature data of LUSC patients.
Figure 1. Volcano maps and heat maps of differentially expressed RNAs between lung squamous cell carcinoma tissues and normal tissues. (A) Volcano plot of LUSC-specific lncRNAs. (B) Volcano plot of LUSC-specific miRNAs. (C) Volcano plot of LUSC-specific mRNAs. (D) The hierarchical clustering heat maps of LUSC-specific lncRNAs. (E)The hierarchical clustering heat maps of LUSC-specific miRNAs. (F) The hierarchical clustering heat maps of LUSC-specific mRNAs.
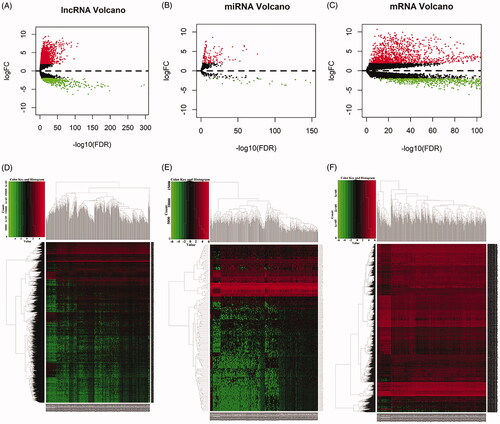
Table 2. The differentially expressed lncRNAs in lung squamous cell carcinoma patients.
Table 3. The differentially expressed mRNAs in lung squamous cell carcinoma patients.
Table 4. The differentially expressed miRNAs in lung squamous cell carcinoma patients.
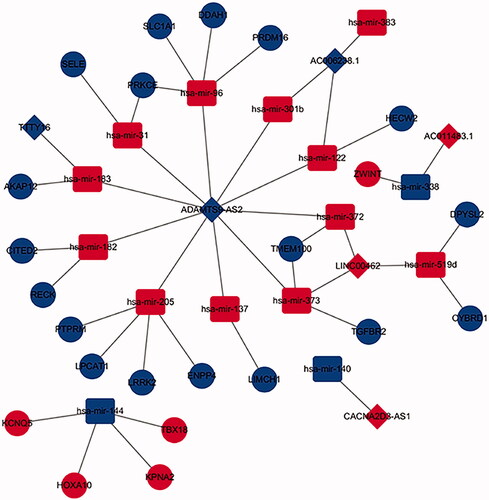
Figure 2. Venn diagram of mRNAs included in ceRNA network and construction of lncRNA-miRNA-mRNA network in LUSC. (A) The red area represents the number of differentially expressed mRNA, the blue area shows the number of targets differentially expressed mRNAs. The purple area in the middle indicates the number of mRNAs which both belonged to the differentially expressed mRNAs and the target mRNAs. (B). The lncRNAs, miRNAs and mRNAs were represented by diamonds, rounded rectangles and ellipses, respectively. The red nodes indicate high expression, and the blue nodes indicate low expression.
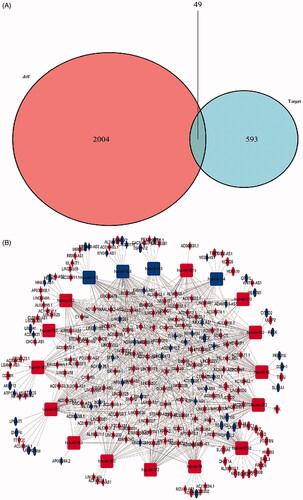
Figure 3. A flow diagram of ceRNA network construction in LUSC. DEmRNAs, DEmiRNAs and DElncRNAs with fold change >2 and p values <0.01 were used; lncRNA-miRNA interaction was predicted by miRcode; lncRNAs that were not associated with DEmiRNAs were removed. mRNAs targeted by miRNA using miRDB, miRTarBase and TargetScan databases; mRNAs that were not DEmRNAs were removed; ceRNA network was constructed.
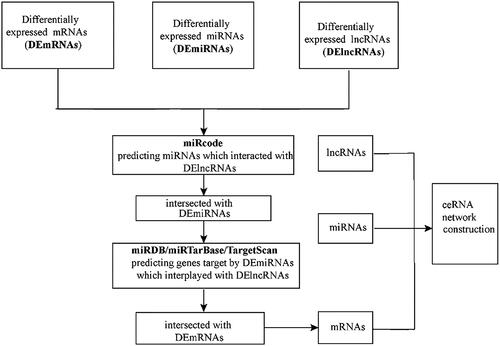
Table 5. Representative relationships between lncRNAs and miRNAs in lung squamous cell carcinoma patients.
Table 6. Representative relationships between miRNAs and mRNAs in lung squamous cell carcinoma.
Figure 4. Kaplan–Meier curve analysis of DElncRNAs, DEmRNAs, DEmiRNAs and overall survival rate in LUSC samples.
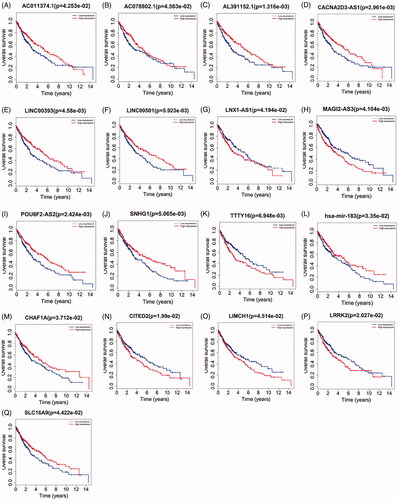
Table 7. Multivariate cox regression analysis of 6 prognostic lncRNAs associated with overall survival in LUSC patients.
Figure 5. Prognostic value of six-lncRNAs in LUSC patients. (A). A risk heat-map established from 6 lncRNAs from 551 LUSC patients. (B). Kaplan-Meier curve analysis for OS (overall survival) of LUSC patients using the 6 lncRNAs signature. (C). ROC curve analysis of the prognostic 6 lncRNAs signature.
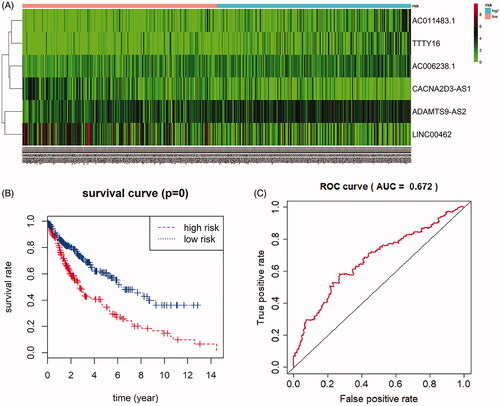
Figure 6. Functional enrichment analysis of lncRNA-mediated differentially expressed mRNAs in LUSC. The outer circle represents the expression (logFC) of DElncRNA-related differentially expressed mRNAs in each enriched GO (gene ontology) terms: red dots which were on the each Go terms indicated the up-regulated differentially expressed mRNAs. Blue dots indicated the down-regulated differentially expressed mRNAs. The inner circle indicates the significance of GO terms (log10-adjusted p values). (A). The circle indicates the correlation between statistically top 30 differentially expressed mRNAs and their gene ontology terms. (B).
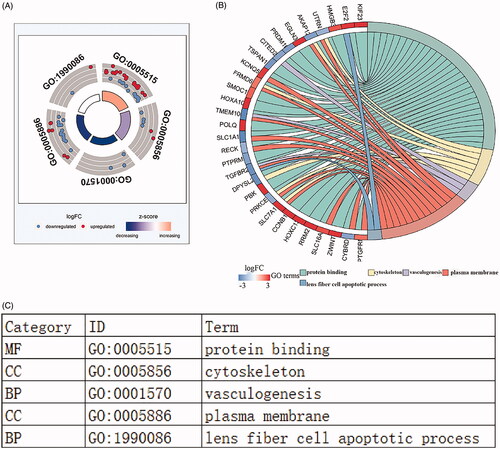
Figure 7. KEGG pathway analysis of differentially expressed mRNAs in LUSC. The top ten pathways lncRNA-related DEmRNAs in barplot and dotplot (A,B). The top 14 pathways were used cytoscape for constructing a pathway-gene network with differentially expressed mRNAs. The purple hubs indicate the pathways, the blue hubs indicate the down-regulated mRNAs, the red hubs indicate the up-regulated mRNAs (C).
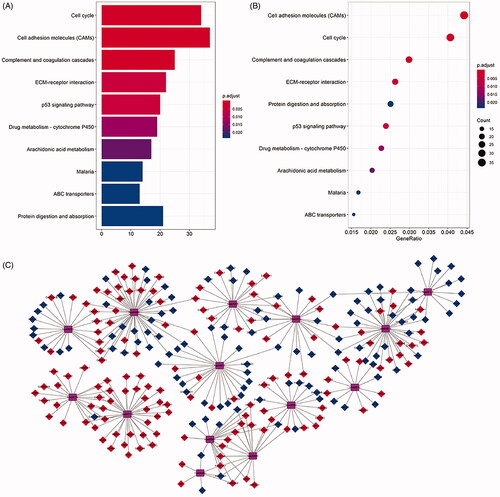
Table 8. The top 14 KEGG pathways enriched by differentially expressed mRNAs.