Figures & data
Figure 1. The expression of lnc-SLC4A1-1 in breast cancer (BC) tissues, blood samples and cell lines. (A) The lnc-SLC4A1-1 level in tumor sample and adjacent normal samples by qRT-PCR. (B) The lnc-SLC4A1-1 level in serum of healthy subjects and BC patients by qRT-PCR. (C) The relationship between lnc-SLC4A1-1 expression and survival times in BC patients. (D) The lnc-SLC4A1-1 level in normal cells (MCF10A) and cancer cells (MDA-MB-231 and MCF-7) by qRT-PCR. *p < .05 and **p < .01 versus Non-tumor, Health, or MCF10A.
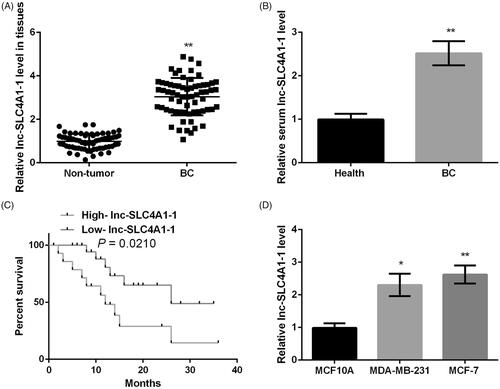
Table 1. Correlation between lnc-SLC4A1-1 expression and clinicopathological characteristics of breast cancer.
Figure 2. Abnormal expression of lnc-SLC4A1-1 was associated with H3K27 acetylation in breast cancer (BC). (A) Example of promoter region of lnc-SLC4A1-1 in UCSC genome browser. (B) The relationship of lnc-SLC4A1-1 expression and acetylation in BC tissues by chromatin immunoprecipitation (ChIP) assay. (C) The relationship of lnc-SLC4A1-1 expression and acetylation in BC cells by ChIP assay. **p < .01 versus Normal or MCF10A.
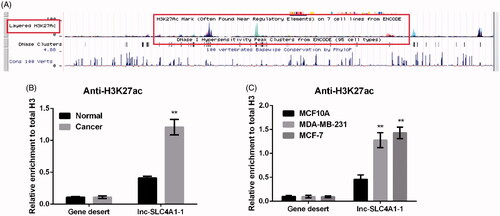
Figure 3. Lnc-SLC4A1-1 knockdown inhibited cell growth in BC cells. (A) lnc-SLC4A1-1 expression of cells transfected with pc-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 or their corresponding control in MDA-MB-231 and MCF-7 cells. (B) Cell viability of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by MTT assay. (C) The number of apoptotic cells of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by flow cytometry analysis. (D) The expression of apoptosis-related proteins (Bcl-2, Bax, pro-caspase-3, cleaved-caspase-3, pro-caspase-9, and cleaved-caspase-9) of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by western blotting. (E) Cell migration of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by Transwell. (F) Cell invasion of untreated cells, cells with sh-NC or sh-lnc-SLC4A1-1 in MDA-MB-231 and MCF-7 cells by Transwell. (G) The expression of epithelial-mesenchymal transition-related proteins (E-cad, N-cad, Vim, and Snail) of untreated cells, cells with TGF-β1, sh-NC + TGF-β1 or sh-lnc-SLC4A1-1 + TGF-β1 in MDA-MB-231 and MCF-7 cells by western blotting. *p < .05, **p < .01 and ***p < .001.
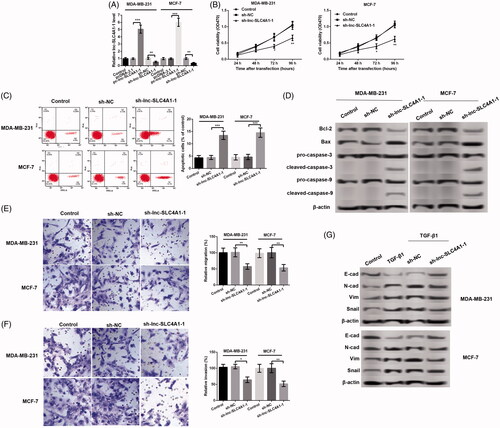
Figure 4. Lnc-SLC4A1-1 promoted CXCL8 expression. (A) Identification of the transcription factors (P50/P105, P65, AP-1 and c-Jun) related to lnc-SLC4A1-1 by RNA immunoprecipitation assay. (B) The expression of NF-κB p65 and CXCL8 in cells treated with pc-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 or pc-lnc-SLC4A1-1 + NF-κB antagonist in MDA-MB-231 cells by western blotting. (C) The expression of NF-κB p65 and CXCL8 in cells treated with pc-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 or pc-lnc-SLC4A1-1 + NF-κB antagonist in MCF-7 cells by western blotting. *p < .05, **p < .01 and ***p < .001.
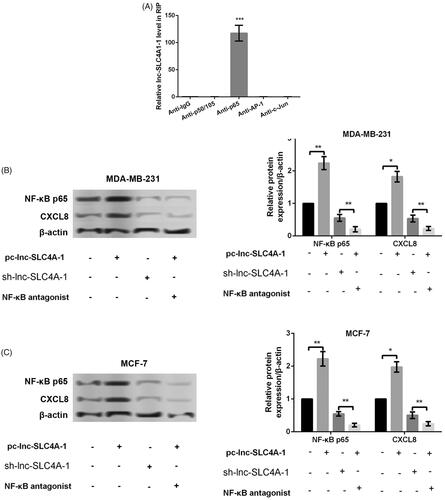
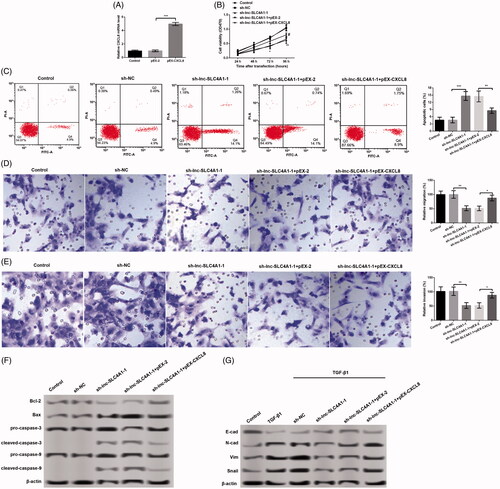
Figure 5. Relationship of CXCL8 and the effects of lnc-SLC4A1-1 in breast cancer. (A) CXCL8 expression in MCF-7 cells transfected with pEX-2, or pEX-CXCL8. (B) Cell viability of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by MTT assay. (C) The number of apoptotic cells of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by flow cytometry analysis. (D) Cell migration of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by Transwell. (E) Cell invasion of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by Transwell. (F) The expression of apoptosis-related proteins (Bcl-2, Bax, pro-caspase-3, cleaved-caspase-3, pro-caspase-9, and cleaved-caspase-9) of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by western blotting. (G) The expression of epithelial-mesenchymal transition-related proteins (E-cad, N-cad, Vim and Snail) of untreated cells, cells with sh-NC, sh-lnc-SLC4A1-1, sh-lnc-SLC4A1-1 + pEX-2, or sh-lnc-SLC4A1-1 + pEX-CXCL8 in MCF-7 cells by western blotting. *p < .05, **p < .01 and ***p < .001.